Challenge #1
Click image to enlarge
PREDICT HITS FOR THE WDR DOMAIN OF LRRK2
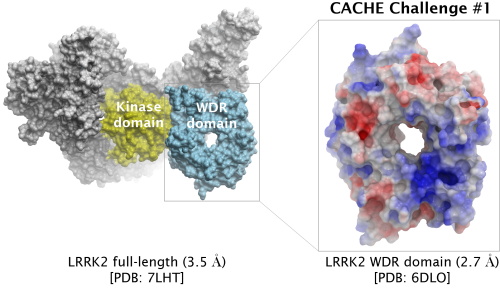
The first CACHE Challenge target is LRRK2, the most commonly mutated gene in familial Parkinson's Disease.
Participants are asked to find hits for the WD40 repeat (WDR) domain of LRRK2. Read more under Details below.
This challenge is supported by the Michael J. Fox Foundation.

CACHE is strongly committed to equity, diversity and inclusion. We strive to create an initiative that welcomes participants from countries, institutions and groups that are underrepresented in our research community. We endeavour to work with individuals who contribute to furthering the diversity of ideas and approaches, welcoming all communities to participate in our challenges.
All applications and submissions to this CACHE Challenge are subject to the CACHE Terms of Participation. Applications should be emailed to [email protected]
2021-12-01
2022-01-30
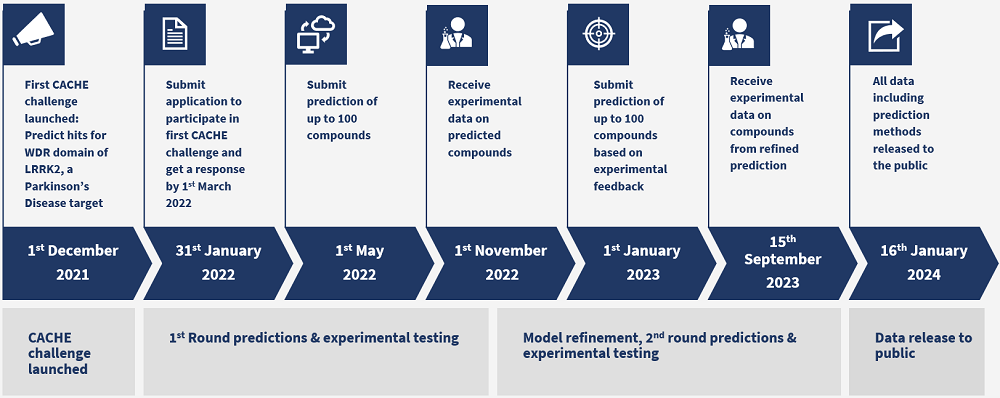
Timeline
Details
Leucine-rich repeat serine/threonine-protein kinase 2 (LRRK2)
| Parameter | Description | |
|---|---|---|
| Gene/UniProt | LRRK2(PARK8)/Q5S007 | |
| Disease association | Parkinson’s Disease (PD) | |
| Challenge | Finding ligands targeting the central cavity of the WD-40 repeat (WDR) domain (aa 2142-2498) | |
| PDB ID (challenge) | 6DLO (2.7 Å) WDR domain | |
| PDB ID (full-length) | 7LHT (3.5 Å) | |
| Pharmacological landscape |
Drug candidates targeting the kinase domain are in preclinical or early-stage clinical trial, including compounds from Genentech, Pfizer, Novartis and Merck. An antisense oligonucleotide that binds the mRNA of the LRRK2 gene and lowers the cellular level of LRRK2 is also in clinical trials. |
|
| CRISPR cell and animal models reported | 1. In Vitro CRISPR/Cas9-Directed Gene Editing to Model LRRK2 G2019S Parkinson’s Disease in Common Marmosets 2. Generation of two LRRK2 homozygous knockout human induced pluripotent stem cell lines using CRISPR/Cas9 |
|
| Why the WDR domain? | PD-associated LRRK2 mutations tend to promote LRRK2 filament formation and enhance LRRK2 interaction with microtubules. Recent structural data reveals that only compounds stabilizing the open form of LRRK2 antagonize the pathogenic formation of LRRK2 filaments in cells, but most kinase inhibitors stabilize the closed form of LRRK2. An alternative and so far overlooked strategy is to pharmacologically target the WDR domain of LRRK2, which is juxtaposed to the kinase domain. The WDR domain in LRRK2 may be important for recruiting LRRK2 signalling partners or for binding to tubulin. WDR domains are disease-associated and druggable. Identifying chemical starting points binding to the WDR domain of LRRK2 is a novel approach to target this protein. | |
| Potential impact | The public release of chemical starting points for an understudied domain of LRRK2 will offer opportunities to target LRRK2 via an allosteric mechanism and make PROTACs to induce its degradation with ligands not directly interfering with the catalytic activity of the target. |
|
