Challenge #2
Click image to enlarge
FINDING LIGANDS TARGETING THE CONSERVED RNA BINDING SITE OF SARS-CoV-2 NSP13
The second CACHE Challenge target is the NSP13 helicase of SARS-CoV-2.
Participants are asked to find hits for the RNA-binding site of NSP13. Read more under Details below.
This challenge is supported by the NIH's AViDD program.
CACHE is strongly committed to equity, diversity and inclusion. We strive to create an initiative that welcomes participants from countries, institutions and groups that are underrepresented in our research community. We endeavour to work with individuals who contribute to furthering the diversity of ideas and approaches, welcoming all communities to participate in our challenges.
All applications and submissions to this CACHE Challenge are subject to the CACHE Terms of Participation.
2022-06-22
2022-09-03
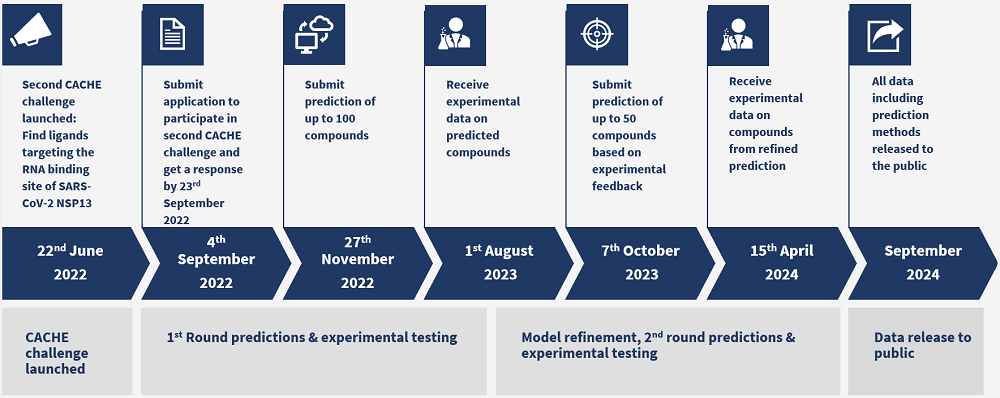
Timeline
Details
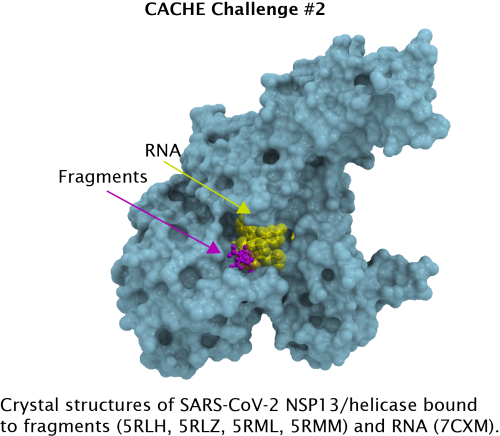
RNA binding site of SARS-CoV-2 NSP13
| Parameter | Description | |
|---|---|---|
| Accession | YP_009725308.1 | |
| Disease association | Alpha- and beta-coronaviruses including SARS-CoV-2 | |
| Challenge | Finding ligands targeting the conserved RNA binding site of SARS-CoV-2 NSP13. Predicted compounds will be procured and tested at CACHE using both enzymatic and binding assays. | |
| Why NSP13? | Helicase inhibition is not a novel anti-viral strategy and potent inhibitors of herpes simplex virus, shingles and hepatitis C viruses have been reported. Helicase inhibitor Pritelivir is in Phase III clinical trials and Amenamevir has been approved in Japan for shingles. In contrast, the SARS-CoV-2 helicase NSP13 has so far been underexplored. The RNA-binding site of NSP13 is the most conserved site in SARS-CoV-2 and across coronaviruses, and the underlying selective pressure is expected to apply to future coronavirus outbreaks: drugs binding this site should be effective against future pandemic threats. RNA unwinding is mechanistically coupled to hydrolysis of a nucleotide triphosphate (generally ATP) at a pocket distinct from the RNA binding site and existing helicase inhibitors either bind the ATP site to prevent ATP hydrolysis, or bind the nucleic acid substrate to antagonize the unwinding activity. The ATP site of NSP13 is significantly less conserved across coronaviruses than the RNA binding pocket . In this CACHE challenge, participants are invited to find ligands that compete with RNA using structure-based drug design (PDB IDs: 5RLH, 5RLZ, 5RML, 5RMM). | |
| Macromolecular structures | The crystal structure of NSP13 has been determined in multiple states and crystal forms. In particular, fragments found in PDB IDs 5RLH, 5RLZ, 5RML, 5RMM overlap with the evolutionary conserved RNA-binding site and may guide the design of more potent molecules. | |
| Pharmacological landscape | There are no drug candidates targeting the SARS-CoV-2 helicase. | |
| Potential impact | The public release of chemical starting points that compete with RNA binding will facilitate the rapid development of selective, potent, cell-active inhibitors. The intended long-term impact is to accelerate the discovery of a coronavirus antiviral that is complementary to existing therapies such as Paxlovid (target: Mpro) and Molnupiravir (target: RdRp) and that would also be effective against future coronavirus outbreaks. | |