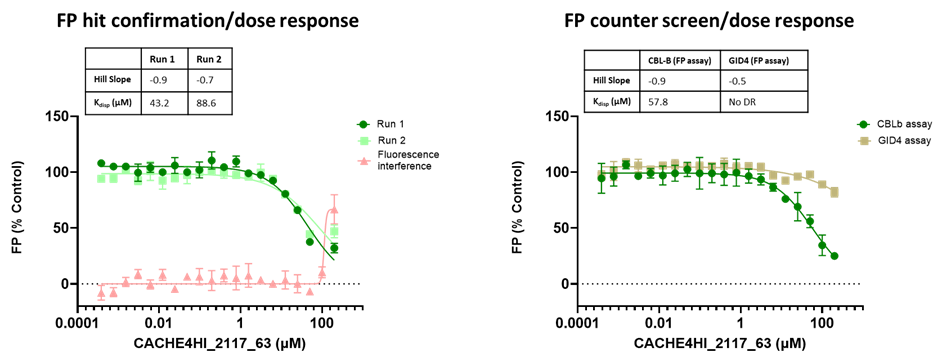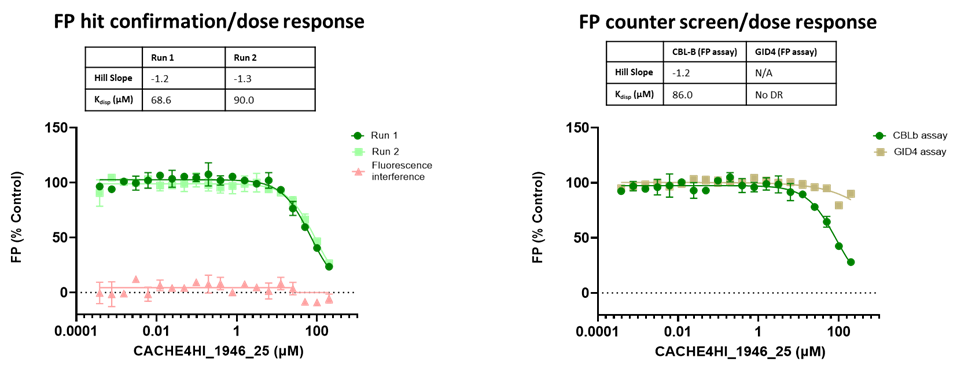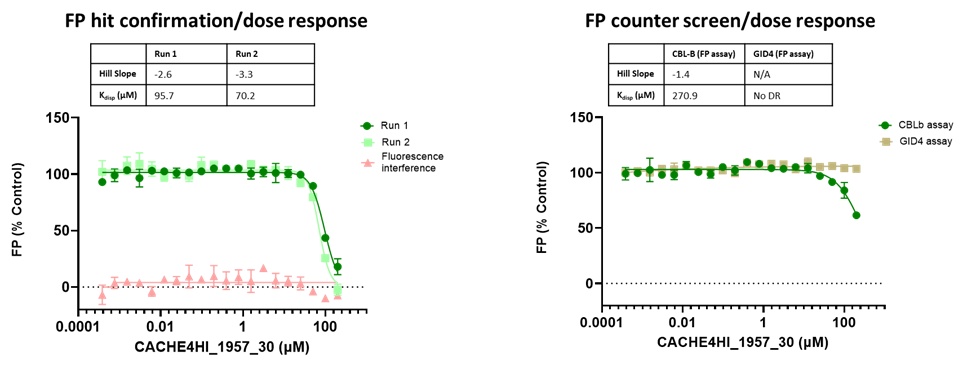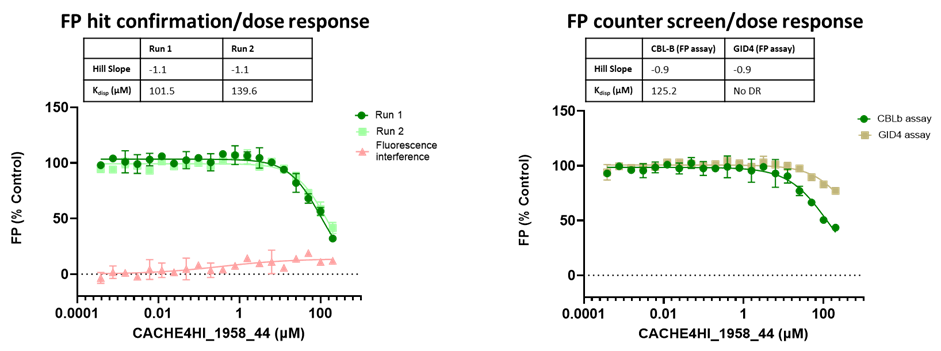Summary of CACHE#4 Round 1 experimental screening pipeline
CACHE#4 compounds are tested experimentally at the Structural Genomics Consortium, University of Toronto, with contributions from Cheryl Arrowsmith, Albina Bolotokova, Renu Chandrasekaran, Irene Chau, Kristina Edfeldt, Pegah Ghiabi, Elisa Gibson, Rachel Harding, Oleksandra Herasymenko, Scott Houliston, Ashley Hutchinson, Mariia Khamina, Peter Loppnau, Matthieu Schapira, Santha Santhakumar and Madhushika Silva.
THE CHALLENGE
- CACHE participants were asked to use their computational methods to find hits for the TKB domain of CBLB. See details.
- After a double-blind peer review where each applicant reviewed 5 applications, 23 participants joined the challenge, representing a diversity of physics-based and AI computational methods.
- Participants collectively selected 1688 compounds (no more than 100 compounds per participant) that we ordered and received from Enamine.
- All experimental data except the structure of the compounds are provided here.
- Chemical structures will be revealed, CACHE participants de-anonymized, and computational methods ranked at the end of the challenge, upon completion of Round 2 (hit expansion).
SUMMARY OF THE EXPERIMENTAL PIPELINE
SUMMARY OF ROUND 1 EXPERIMENTAL RESULTS PER PARTICIPANT
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
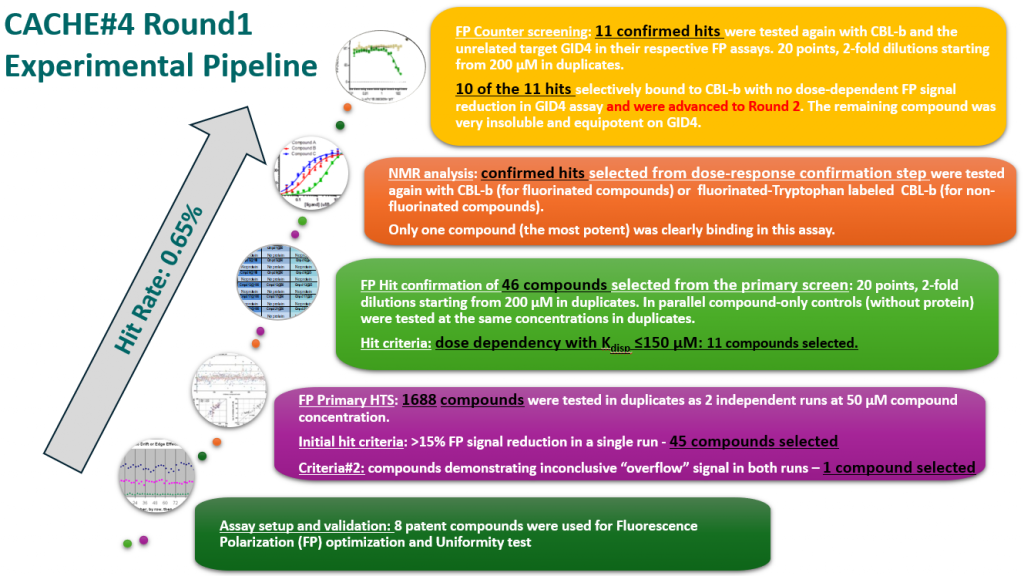
EXPERIMENTAL DETAILS
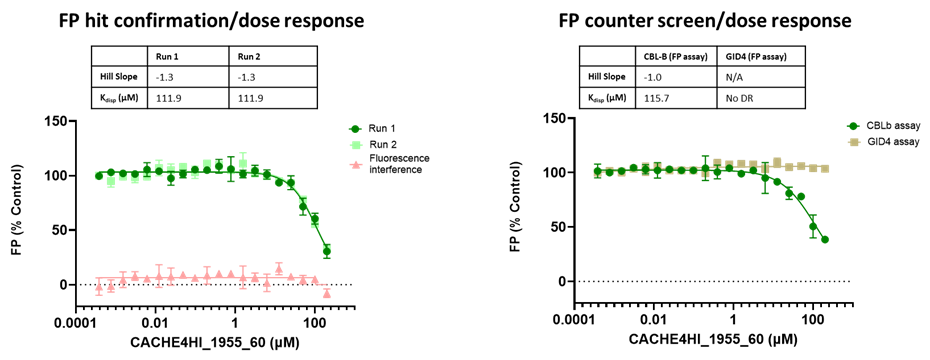
All 1688 compounds were screened in two independent runs at 50 µM in a fluorescence polarization (FP) assay which measured displacement of the fluorescently labelled version of the known CBLB ligand C2787102.
FP probe:
FP Assay conditions: recombinant CBLB [construct 34-427 with C-terminal Avi and N-terminal His tagged] at 625 nM; BODIPY-labeled C2787102 at 10 nM; Buffer: 20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, 0.01% (v/v) TritionX-100, 2% (v/v) DMSO.
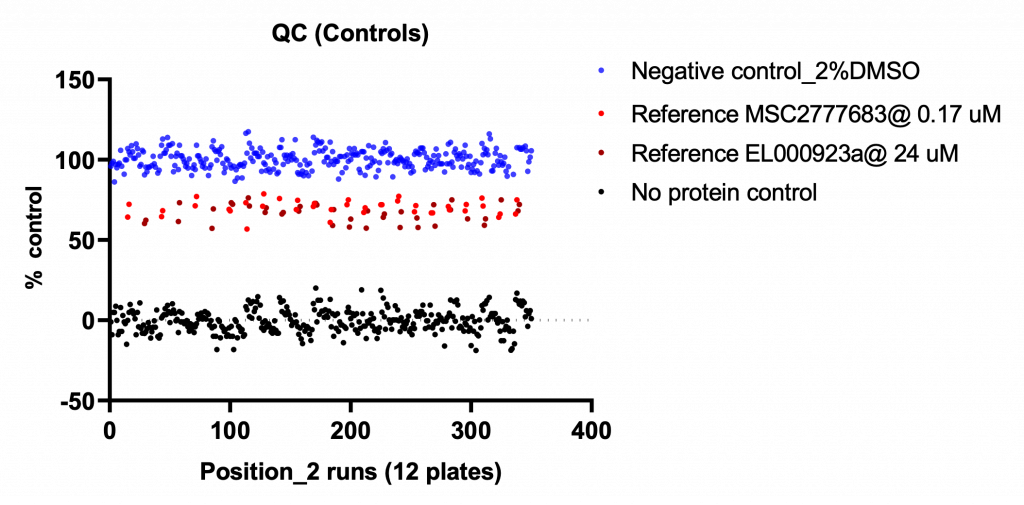
The assay was validated with two positive controls, C2777683 and EL000923. The better Hill Slope of EL000923 reflects the fact that assay conditions were optimized for low affinity compounds.

The assay was quality-controlled and robust with Z´ (0.5-0.7) and S/B (2.1-2.2):
A good correlation was observed between the two independent runs:
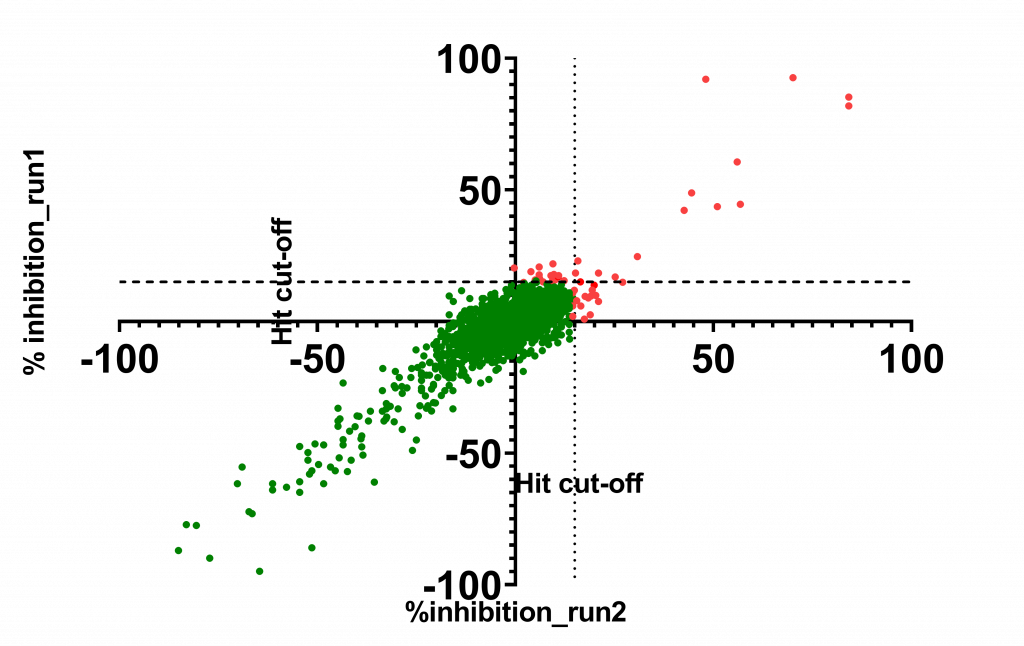
45 compounds with ≥15% inhibition (highlighted in red) in at least one of the two runs were advanced to the dose-response stage. One compound with an overflowed signal was also advanced.
The solubility and aggregation of all 46 compounds of interest was assessed by dynamic light scattering (DLS), showing that several compounds had poor solubility and aggregation properties when solubilized at high concentration in assay buffer (DLS data is included in the excel file and explained here).
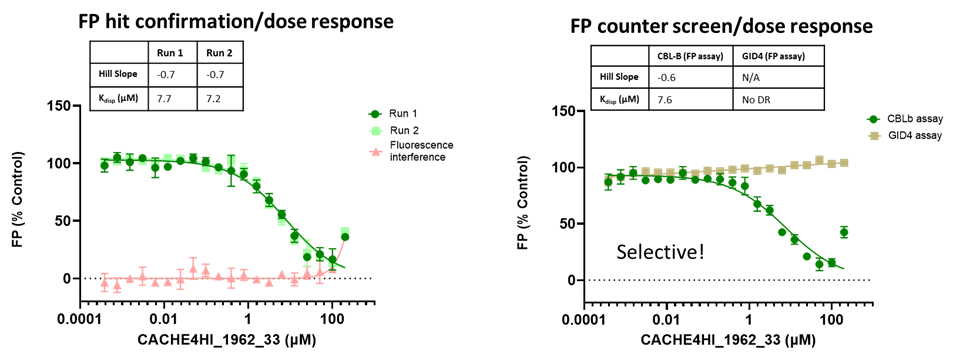
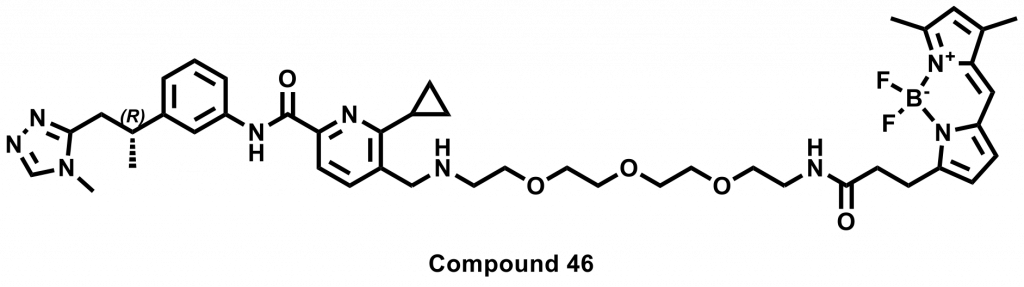
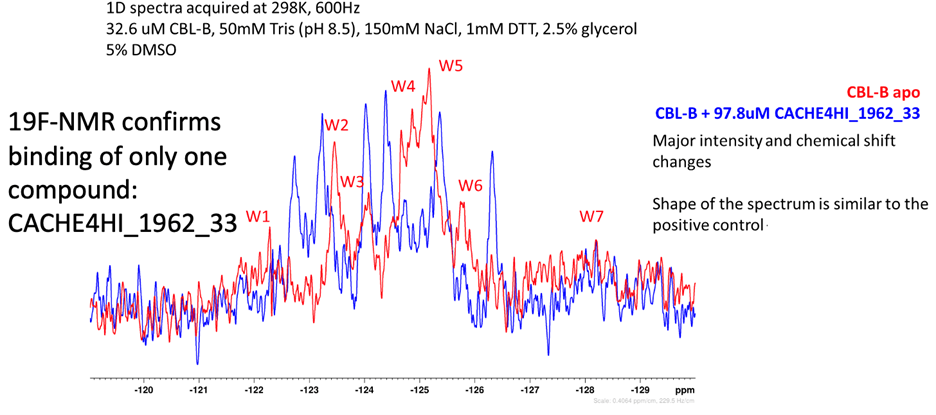
Eleven of the 46 compounds had a measurable Kdisp < 150 µM and their binding was tested in a 19F-NMR assay. Non-fluorinated compounds were tested with a Trp-fluorinated version of CBLB. Only one compound, CACHE4HI_1962_33, was clearly binding to CBLB in this assay. It should be noted that (1) this was the most potent of the 11 compounds of interest and (2) unlike other compounds, this is a close analog of patent compounds, diminishing the value of this hit.
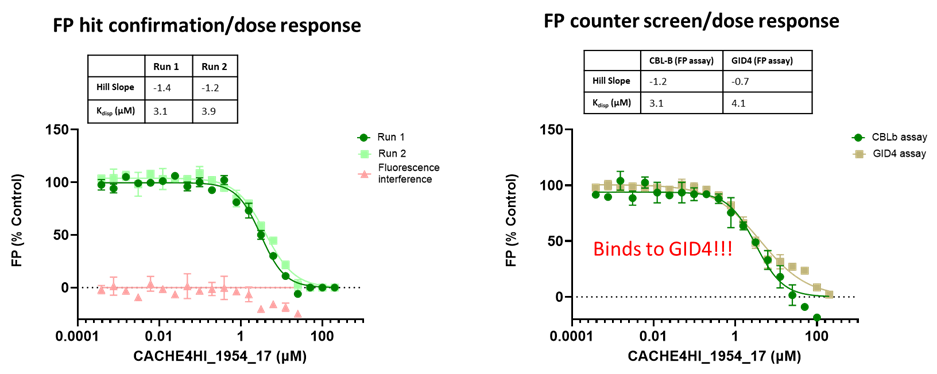
The promiscuity of all eleven compounds of interest was assessed by testing their binding to an unrelated target, GID4, in a pre-established FP assay where GID4 chemical probe PFI-7 was used as a positive control:
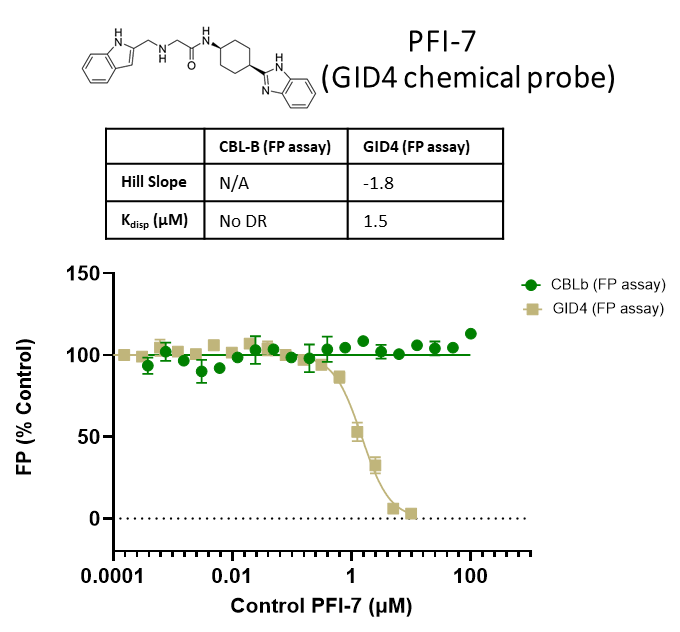
One compound with poor solubility, CACHE4HI_1954_17, was equipotent in the CBLB and GID4 FP assays and dismissed. Ten other compounds were selective for CBLB and advanced to Round 2: hit expansion. Some of these compounds are weak and/or tend to aggregate at high concentration in the absence of detergent but were still advanced to Round 2 to avoid false negatives. The aim of Round 2 is to generate convincing SAR around these hits and flag false positives. We hope analogs with increased potency will show a binding signal in the 19F NMR assay.
FP dose response curves of 11 compounds of interest showing specific binding to CBLB versus GID4 for 10 compounds: