Results of CACHE Challenge #3
CACHE#3 participants who asked to be de-anonymized:
- Rahul Ravichandran, Francesco Gentile (University of Ottawa), Fuqiang Ban, Artem Cherkasov (University of British Columbia).
- Marko Breznik, Valerij Talagayev, Yu Chen, Sijie Liu , Amit Pandit, Clemens Alexander Wolf, Niklas Piet Doering, Katrin Denzinger, Dr. Kristina Puls, Gerhard Wolber(Freie Universität Berlin).
- Minghu Song, Xinyu Wang, Xutao Shi (University of Connecticut), Kang Ding, Bo Huang, Huting Wang, Boyang Jia, Hongbo Zhang (Beijing StoneWise Technology Co).
- Charuvaka Muvva, Durai Prasanna Venkatesh, Keuwan Park (Korea Institute of Science and Technology).
- Jude Wells, Dénes Berta, Wim Dehaen, Andrea Karlova, Sam Martino, Brooks Paige, Neeladri Sen, Geoff Wells, Sarah Harris, Daniel Felfoldi, Christine Orengo, Edina Rosta (University College London).
- Fold- it citizen scientists, Rocco Moretti,Thomas Scott, Jens Meiler (Vanderbilt University).
- Aziz Abu-Saleh, Purshotam Sharm, John Trant (University of Windsor).
- Alexander Tropsha, Konstantin Popov ( UNC Chapel Hill) & Dmytro Kozakov ( Stony Brook University)
- Laurent Hoffer, Kevin Bishop, Hugo Bohorquez, David Uehling, Rima Al-awar, Gennady Poda (Ontario Institute for Cancer Research).
- Shuangjia Zheng (Sun Yat-sen University).
- Francois Sindt, Didier Rognan (Université de Strasbourg).
- Shubhangi Kandwal, Darren Fayne (Dublin City University).
- Damon Tingey, Eliza Kartchner, Denis Dell Corte ( Brigham Young University).
- Casper Steinmann, Jan Jensen (University of Copenhagen).
- Mykola Protopopov, Yurii Moroz ,Olga Tarkhanova (Chemspace LLC).
- Juyong Lee (Seoul National University).
- Ozan Gokdemir, Xuefeng Liu, Austin Clyde (University of Chicago).
CACHE#3 compounds were tested experimentally at the Structural Genomics Consortium, University of Toronto, with contributions from Suzanne Ackloo, Cheryl Arrowsmith, Albina Bolotokova, Irene Chau, Kristina Edfeldt, Elisa Gibson, Oleksandra Herasymenko, Rachel Harding, Ashley Hutchinson, Peter Loppnau, Matthieu Schapira, Almagul Seitova and Madhushika Silva. Structural biology was conducted by Galen Correy at UCSF, in the lab of James Fraser, who served as Target Expert for CACHE #3.
SUMMARY
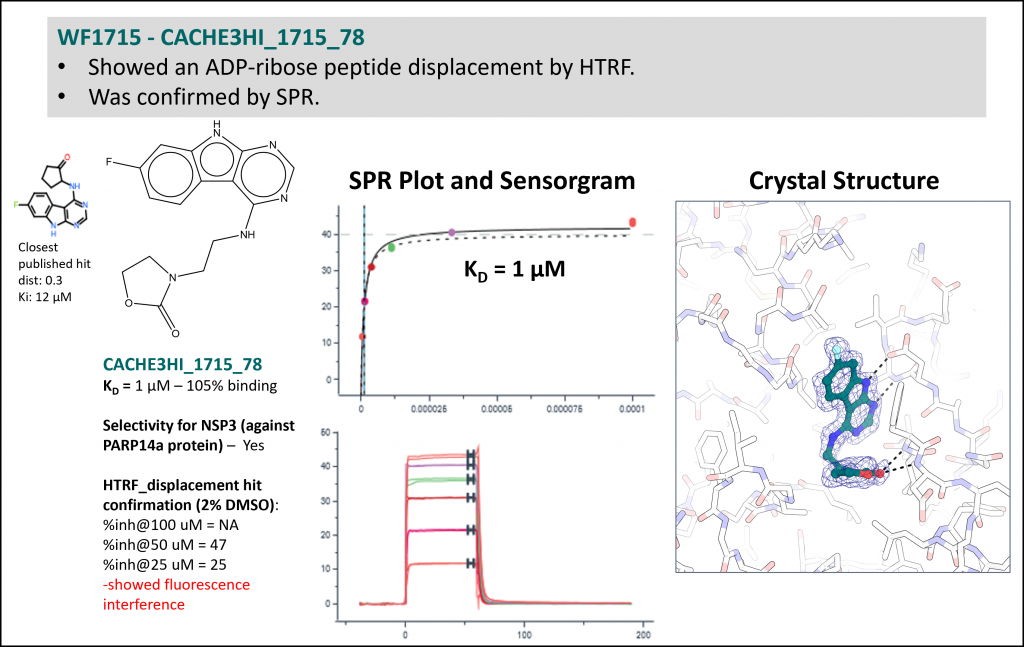
23 computational teams selected up to 100 compounds each. The resulting 1739 compounds were tested experimentally. After a hit expansion round, teams were ranked based on the activity and novelty of their predicted molecules. Results are summarized in Table 1 below. The first 4 teams each had one experimentally confirmed compound that was sufficiently novel (binding affinities ranging from 15 to 100 µM). Other teams (ranks 5 to 9) had one or more confirmed hits (binding affinities starting at 1 µM - CACHE3HI_1715_78) but they were close analogs of hits previously published in PNAS. Computational workflows used by these 9 teams are schematized in Box 1 below. Compounds are shown at the end of this page. In total, 2039 CACHE compounds were tested experimentally at the Structural Genomics Consortium, and the crystal structures of 44 were solved in complex with Nsp3 Mac1 in James Fraser's lab at UCSF.
INTRODUCTION
- For CACHE #3, 23 participants from 11 countries used their computational workflows to predict small molecule ligands that bind to the ADP-ribose binding site of the Macrodomain 1 of SARS-CoV-2 NSP3, a domain used by the virus to evade the host immune system. At the start of this challenge, multiple ligands had been reported in a PNAS paper with multiple structures in the PDB. The most potent molecules in PNAS had a carboxylic acid, which was identified as chemical liability.
- CACHE Challenges begin with a hit finding round (Round 1), where participants can nominate up to 100 commercially available compounds. Round 1 predictions are then experimentally tested and the resulting data returned to participants. In the hit-expansion round (Round 2), participants whose nominated compounds show some sign of activity in experimental binding assays (compounds of interest) can select up to 50 follow-up compounds. Together, these two rounds are designed to avoid both false positives and false negatives, which is critical to evaluate computational methods properly.
- In a separate exercise, computational teams are asked to predict hits from a library composed of the merged Round 1 selections from all participants. Here, all teams are screening the same library, and all compounds are experimentally characterized as active or inactive, but experimental data is blinded to participants.
THE CHALLENGE
- CACHE participants were asked to predict compounds with novel chemical templates without carboxylic acids and that compete with the substrate, ADP-ribose (see details)
- After a double-blind peer review where each applicant reviewed 5 applications, 23 participants joined the challenge, representing a diverse array of physics-based and AI computational methods.
- Participants collectively selected 1739 compounds (no more than 100 compounds per participant) that were ordered from Enamine and tested experimentally.
ROUND 1 RESULTS
All Round 1 experimental data are provided here.
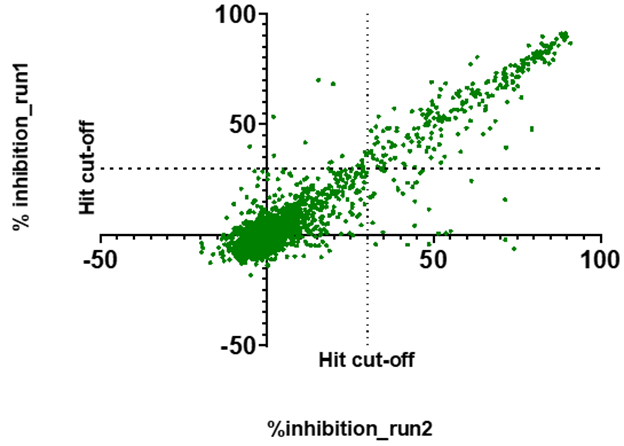
- All 1739 compounds were screened in two independent runs at 100 µM in a fluorescence-based HTRF competition assay as described in https://doi.org/10.1073/pnas.2212931120
- An excellent correlation was observed between the two runs:
- Hit candidates were tested in a dose-response experiment.
- The aggregation and solubility of hit candidates was measured by dynamic light scattering (DLS) as described here and in this short introduction.
- 281 compounds showed >30% inhibition in at least one of the runs and were advanced to the next step. An additional 21 compounds showed inconclusive signal (overflow) in both runs and were also advanced to avoid false negatives.
- The resulting 302 compounds were re-tested in the HTFR assay at 25, 50 and 100 µM in duplicate. 150 compounds showed clear dose dependent inhibition, >30% inhibition at 100 µM, no fluorescence interference and were advanced to the next step. The signal was ambiguous for another 16 compounds that were also advanced to the next step, to avoid false negatives.
- The solubility and aggregation of 166 compounds was measured by dynamic light scattering (DLS) as described here and in this short introduction to determine the maximum concentration that could be used in the subsequent SPR dose response experiment.
- An SPR binding assay was validated with ADP-ribose and three positive controls, published in https://doi.org/10.1073/pnas.2212931120
Z5010894404 published Ki: 0.4 µM
Z5265428218 published Ki: 1.5 µM
Z5010894395 published Ki: 24 µM
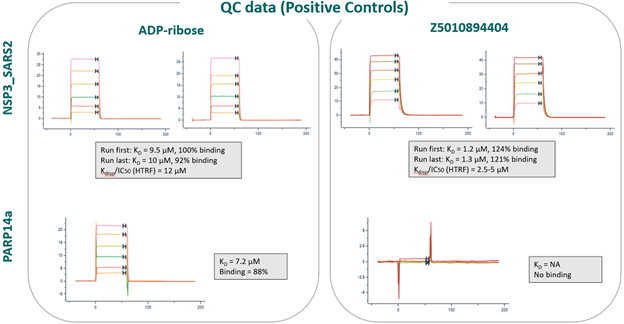
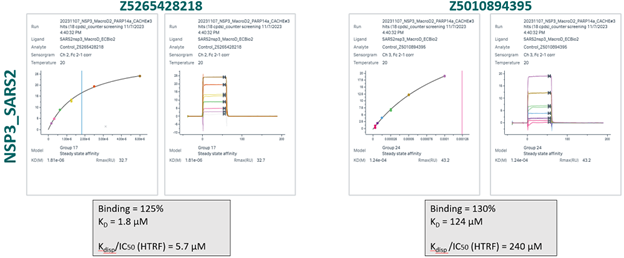
- 166 compounds were tested in a 6-point dose-response experiment by SPR. Top concentration was set to 100 µM, 50 µM or 30 µM, based on the solubility measured by DLS. 30 compounds had KD < 100 µM, > 30% of expected binding level, Chi2 < 10% Rmax and were advanced to the next step. Another 6 compounds had unclear data and were also advanced.
- 36 compounds were tested again by SPR, but with 4% DMSO instead of 2% DMSO to further increase the solubility of the compounds. Compounds were tested both against NSP3 (target) and PARP14 (anti-target), but PARP14 data was not used to dismiss compounds. 28 compounds of interest selected by 9 participants were confirmed and advanced to Round 2: hit expansion. These 9 participants were asked to select 50 follow-up compounds to generate a convincing SAR around their experimental hits.
- All SPR data, along with SPR sensorgrams, are available at https://public.thesgc.org/hitconfirmation/spr.php
- The structure of 44 CACHE compounds bound to Nsp3-Mac1 was solved by X-ray crystallography. Detailed data are available here.
ROUND 2 RESULTS
All Round 2 experimental data are provided here.
- 296 analogs of the 28 compounds of interest advanced to Round 2 were selected by 9 participants to further build experimental support for their computational predictions.
- As in Round 1, the experimental pipeline was HTRF followed by SPR.
- All 296 compounds were screened by HTRF. 114 compounds, including 17 re-supplied parent hit molecules, showed clear dose dependent inhibition, >30% inhibition at any tested concentration, no fluorescence interference and were advanced to the next step. Additionally, 33 compounds that showed >30% inhibition at 100 µM with no dose dependent inhibition or >20% inhibition at 100 µM with a dose dependent inhibition were also advanced.
- The maximum compound concentration that could be used in the following SPR dose response experiment was based on compound solubility evaluated by DLS.
- 158 compounds selected above, in addition to 16 Round 1 compounds that produced signal artifact in HTRF but were rescued by X-ray crystallography, were tested in a 6-points dose-response by SPR.
- 84 Round 2 analogs showed a dose-dependent displacement of ADP-ribose peptide by HTRF and confirmed binding by SPR.
- All hit compounds were tested against NSP3 (target) and PARP14 (anti-target) to verify that binding was specific.
- All SPR data, along with SPR sensorgrams, are available at https://public.thesgc.org/hitconfirmation/spr.php. Detailed slides on CACHE 3 hits are available here.
EVALUATION OF COMPUTATIONAL METHODS
- The biophysical data and SAR of Round 1 hits and their Round 2 follow-ups were evaluated by an independent Hit Evaluation Committee composed of industry experts in biophysics (Anders Gunnarsson, Astra Zeneca & Vera Puetter, Nuvisan), medicinal chemistry (Hartmut Beck, Bayer & Lars Wortmann, Boehringer Ingelheim), and computational chemistry (Pat Walters, Relay Therapeutics ) leading to a final score assigned to each Round 1 hit.
- In addition to the experimental binding data, the Committee evaluated any chemical liability, the chemical novelty compared with compounds published in the PNAS paper (the only paper that was available before participants made their predictions) and the presence of a carboxylic acid (participants were asked to exclude this functional group at the outset of this challenge).
- The results are provided Table 1. Interestingly, while the vast majority of Round 1 compounds did not have one of the two chemical scaffolds found in most molecules published in PNAS, almost none of these novel compounds were confirmed experimentally. Five teams (Ranks 5 to 9) had some predictions confirmed experimentally, but compounds were not judged novel. Four teams (Ranks 1 to 4) predicted one chemically novel compound that was subsequently confirmed experimentally.
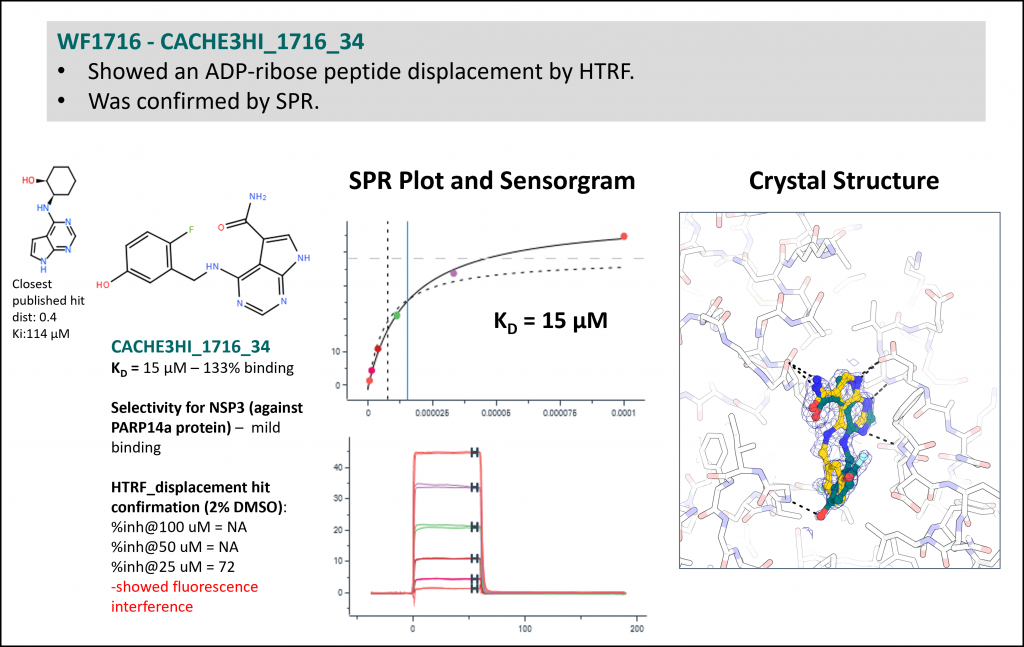
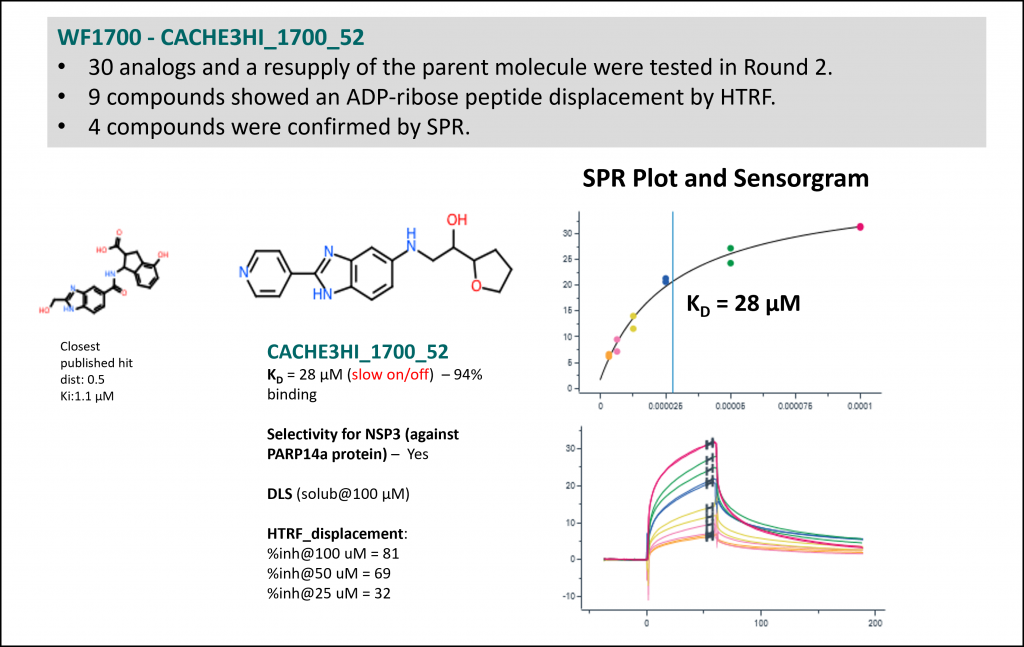
- Workflow 1700 produced the most chemically novel hit. However, SPR data was not reproducible with a resupplied batch in Round 2 (HTRF data was). Additionally, Round 2 analogs were not closely related. This is why this workflow is not ranked at the top in Table 1.
- The structure, SAR and closest published analog of all hits are provided here.
| Rank | Workflow | Participant | Compounds Tested | Hits | Chemically Novel Hits | Top Score | Aggregated Score | Top Novelty Score | Carboxylic Acid Penalty |
| 1 | 1706 | Francesco Gentile [U Ottawa], Artem Cherkasov [U British Columbia] | 83 | 1 | 1 | 21.5 | 21.5 | 2 | |
| 2 | 1705 | Gerhard Wolber [Freie universitat Berlin] | 66 | 1 | 1 | 19.4 | 19.4 | 2 | |
| 3 | 1716 | Minghu Song [U Connecticut] | 51 | 1 | 1 | 18.1 | 18.1 | 2 | |
| 4 | 1700 | Keunwan Park [Korea Institute of Science and Technology] | 73 | 1 | 1 | 21.3 | 21.3 | 3 | Yes |
| 5 | 1715 | Edina Rosta [University College London] | 92 | 11 | 0 | 23.8 | 220.6 | 0 | |
| 6 | 1690 | Rocco Moretti, Jen Meiler [Drug-it crowd-sourcing] | 82 | 6 | 0 | 23 | 126 | 0 | |
| 7 | 1696 | John Trant [Binary Star Research Services, Canada] | 64 | 4 | 0 | 23.5 | 87.4 | 0 | |
| 8 | 1718 | Tropsha [UNC], Kozakov [ Stony Brook University | 86 | 2 | 0 | 23.1 | 46.3 | 0 | |
| 9 | 1708 | Gennady Poda [Ontario Inst. Cancer Centre] | 96 | 2 | 0 | 21.9 | 38.7 | 0 | |
| 10 | 1714 | 41 | 1 | 1 | 12.9 | 12.9 | 3 | ||
| 11 | 1709 | Shuangjia Zheng [Sun Yat-sen University] | 59 | 1 | 0 | 9.4 | 9.4 | 0 | |
| 12 | 1687 | Didier Rognan [CNRS] | 61 | 0 | |||||
| 13 | 1689 | Darren Fayne [Trinity College Dublin] | 89 | 0 | |||||
| 14 | 1691 | Dennis Della Corte [Brigham Young University] | 71 | 0 | |||||
| 15 | 1694 | Jan Jensen [University of Copenhagen] | 83 | 0 | |||||
| 16 | 1695 | 99 | 0 | ||||||
| 17 | 1697 | Olga Tarkhanova [Chemspace LLC] | 66 | 0 | |||||
| 18 | 1702 | 91 | 0 | ||||||
| 19 | 1704 | Juyong Lee [Seoul National University] | 89 | 0 | |||||
| 20 | 1707 | 97 | 0 | ||||||
| 21 | 1712 | 73 | 0 | ||||||
| 22 | 1717 | 82 | 0 | ||||||
| 23 | 1879 | 45 | 0 | ||||||
Table 1: Ranked teams and links to computational workflows; Compounds tested: number of compounds tested in Round 1; Hits: number of compounds of interest advanced to Round 2; Chemically Novel Hits: number of chemically novel Round 1 hits; Top score: score from the Hit Evaluation Committee of the best Round 1 hit; Aggregated score: cumulative score of all Round 1 hits. Top novelty score: chemical novelty score of the most novel compound [the score indicates the number of medicinal chemists who judged a compound chemically novel]. Carboxylic penalty: many analogs in Round 2 had a carboxylic acid.
- In a separate evaluation scheme, all 1739 compounds selected in Round 1 were merged and participants were asked to predict active molecules from this merged library. This is a complementary exercise as here, all participants screen the same set of compounds. To evaluate the predictions, experimentally confirmed hits with a score from the Hit Evaluation Committee greater than 15 were clustered into two tiers: four “Tier 1” compounds that were chemically novel and 25 “Tier 2” compounds judged not chemical novel by the Committee. The results are summarized in Figure 1 and detailed here.
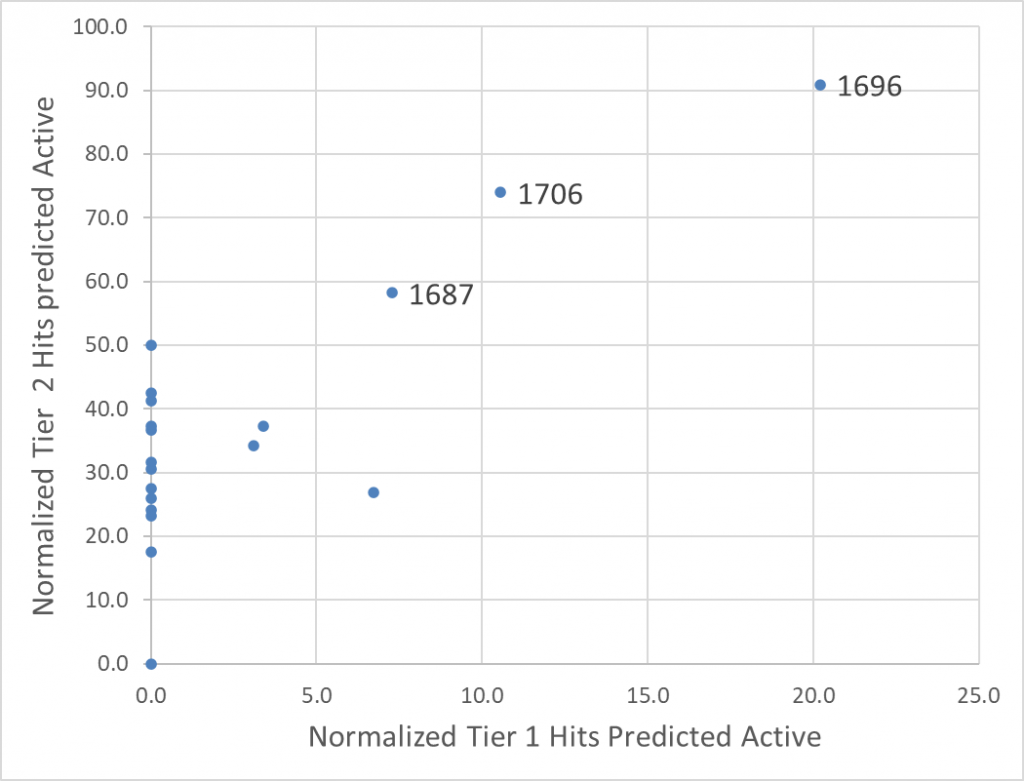
Figure 1: Normalized number of experimentally confirmed Tier 1 hits (chemically novel) and Tier 2 hits (not novel) predicted active when screening all 1739 Round1 compounds. Three groups stand out in this exercise. Details available here.
COMPUTATIONAL METHODS
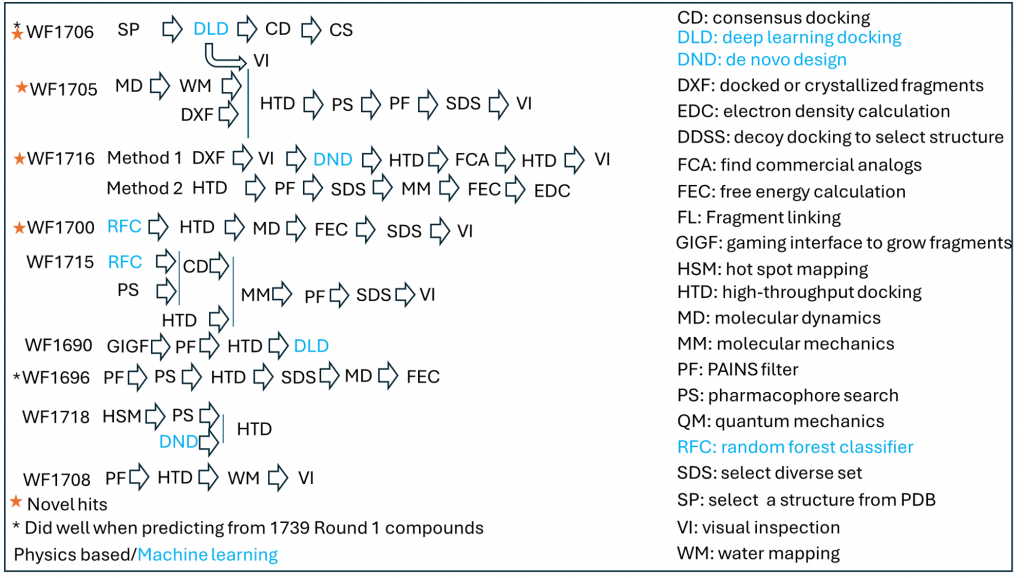
Box 1: Schematics of the best performing computational methods.
NOVEL NSP3-MAC1 LIGANDS
Experimental data for the top four compounds (Rank 1 to 4 Table 1) are shown below.
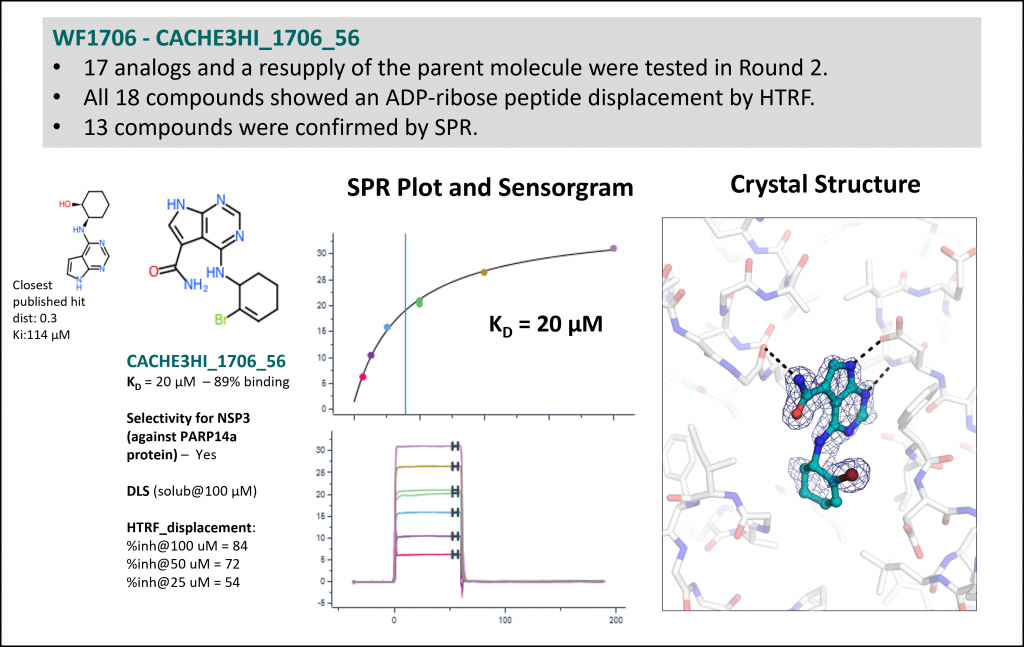
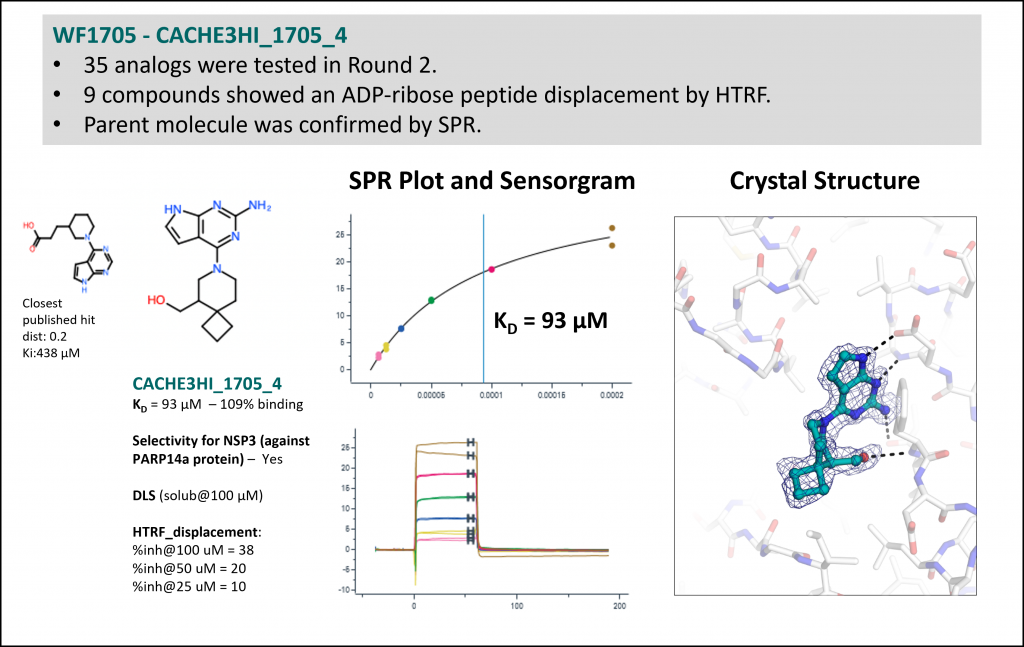
Experimental data for the most potent but chemically less novel compound (one of 11 hits from workflow 1715: Rank 5, Table 1)