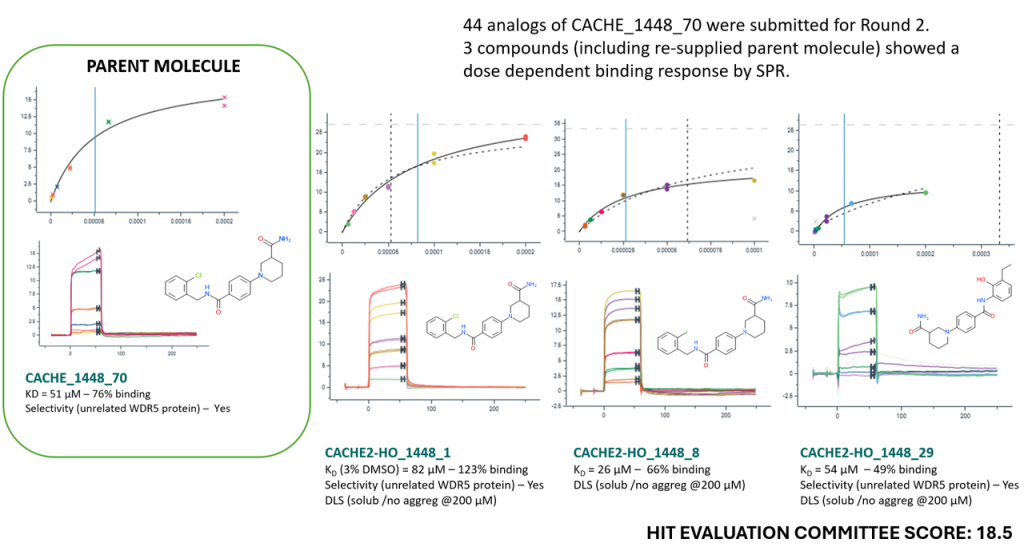
Results of CACHE Challenge #2
CACHE#2 participants who asked to be de-anonymized:
- Karina dos Santos Machado, Adriano Velasque Werhli (Universidade Federal do Rio Grande - FURG, Rio Grande, Brazil), Frederico Schmitt Kremer (OmixLab, Universidade Federal de Pelotas, Capão do Leão, Brazil)
- David Koes, Ian Dunn (University of Pittsburgh, Pittsburgh, U.S.)
- Dmitri Kireev, Akhila Mettu, Xiaowen Wang (University of Missouri, St. Louis, U.S.)
- Rocco Moretti, Thomas Scott (Vanderbilt University, Nashville, U.S.), Jens Meiler (Vanderbilt University, U.S. & Leipzig University, Germany), Foldit Players
- Yurii Moroz, Olga Tarkhanova, Mykola Protopopov (Chemspace, Kyiv, Ukraine)
- Gennady Poda, Laurent Hoffer (Ontario Institute for Cancer Research and University of Toronto, Canada)
- Vincent Blay, Miles McGibbon, Sam Money-Kyrle, Douglas R. Houston (University of Edinburgh, Scotland)
- Ben Cree, Rachael Pirie, Mateusz Bieniek, Joshua Horton, Roly Armstrong, Kate Harris, Natalie Tatum, Daniel Cole (Newcastle University, U.K.)
- Keunwan Park and Prasannavenkatesh Durai (Korea Institute of Science and Technology, Republic of Korea)
- Zheng Zhao, Philip E. Bourne (University of Virginia, Charlottesville, U.S.)
- Shuangjia Zheng (Shanghai Jiao Tong University, Shanghai, China), Wei Lu and Jixian (Aureka Bio)
- Jan Halborg Jensen (University of Copenhagen, Denmark) and Casper Steinmann (Aalborg University, Denmark)
- Shubhangi Kandwal (Dublin City University and Trinity College Dublin, Ireland) and Darren Fayne (Dublin City University, Ireland)
- Marko Breznik, Sijie Liu, Katrin Denzinger, Amit Pandit, Leon Obendorf, Valerij Talagayev, Gerhard Wolber (Free University of Berlin, Berlin, Germany), Federico Ricci (University of Messina, Messina, Italy)
- Emanuele Carosati (University of Trieste, Italy), Marko Jukič (University of Maribor, Slovenia)
- Jude Wells, Edina Rosta, (University College London, London, U.K.)
CACHE#2 compounds were tested experimentally at the Structural Genomics Consortium, University of Toronto, with contributions from Cheryl Arrowsmith, Albina Bolotokova, Peter J. Brown, Irene Chau, Kristina Edfeldt, Pegah Ghiabi, Elisa Gibson, Oleksandra Herasymenko, Rachel Harding, Zarah Hejazi, Scott Houliston, Ashley Hutchinson, Carla Kharadjian, Helen Li, Peter Loppnau, Sumera Perveen, Matthieu Schapira, Almagul Seitova and Madhushika Silva. Structural biology was conducted at the Centre for Medicines Discovery, University of Oxford, with contributions from Joseph Newman.
-------------------------------------------------------------------------------------------------------
For CACHE Challenge #2, 23 participants applied their computational methods to predict which small molecule ligands bind to the RNA groove of the SARS-CoV-2 helicase NSP13. Structures in complex with fragments were available at the start of the challenge (PDB codes 5RLH, 5RLZ, 5RML, 5RMM).
CACHE Challenges begin with a hit finding round (Round 1), where participants can nominate up to 100 commercially available compounds. Round 1 predictions are then experimentally tested and the resulting data returned to participants. In the hit-expansion round (Round 2), participants whose nominated compounds show some sign of activity in experimental binding assays (compounds of interest) can select up to 50 follow-up compounds.
Together, these two rounds are designed to avoid both false positives and false negatives, which is critical to evaluate computational methods properly.
THE CHALLENGE
- CACHE participants were asked to use their computational methods to find hits for the RNA-binding site of the SARS-CoV-2 helicase NSP13. See details.
- After a double-blind peer review where each applicant reviewed 5 applications, 23 participants joined the challenge, representing a diverse array of physics-based and AI computational methods.
- Participants collectively selected 1957 compounds (no more than 100 compounds per participant) that were ordered and received from Enamine.
ROUND 1 RESULTS
All Round 1 experimental data are provided here. A summary is available here. In brief:
- 1957 compounds were screened at 50 µM against NSP13 in an SPR binding assay. An introduction on SPR data for non-experts is available here.
- Hit candidates were tested in a dose-response experiment.
- The aggregation and solubility of hit candidates was measured by dynamic light scattering (DLS) as described here and in this short introduction.
- Forty-six compounds of interest selected from 18 participants advanced to the hit-expansion phase (Round 2), where participants selected up to 50 follow-up compounds for experimental characterization.
ROUND 2 RESULTS
All Round 2 experimental data are provided in this excel file and these powerpoint slides.
- 618 compounds from 17 participants were screened at 50 µM in an SPR binding assay followed by dose-response, and measurement of aggregation and solubility, as in Round 1.
- Compounds were also tested in an NSP13 ATPase assay. No correlation was observed with binding by SPR (indeed, it is not established in the literature that compounds binding the RNA groove of NSP13 inhibit the NSP13 ATPase activity coupled to RNA unwinding). The data are provided but was not used to evaluate compounds.
- 19F-NMR was used as an orthogonal binding assay for fluorinated molecules, and results are included in the final powerpoint slides.
EVALUATION OF COMPUTATIONAL METHODS
The biophysical data and SAR of Round 1 hits and their Round 2 follow-ups were evaluated by an independent Hit Evaluation Committee composed of industry experts in biophysics (Anders Gunnarsson, Astra Zeneca & Vera Puetter, Nuvisan), medicinal chemistry (Hartmut Beck, Bayer & Lars Wortmann, Boehringer Ingelheim), and computational chemistry (Pat Walters, Relay Therapeutics ) leading to a final score assigned to each Round 1 hit.
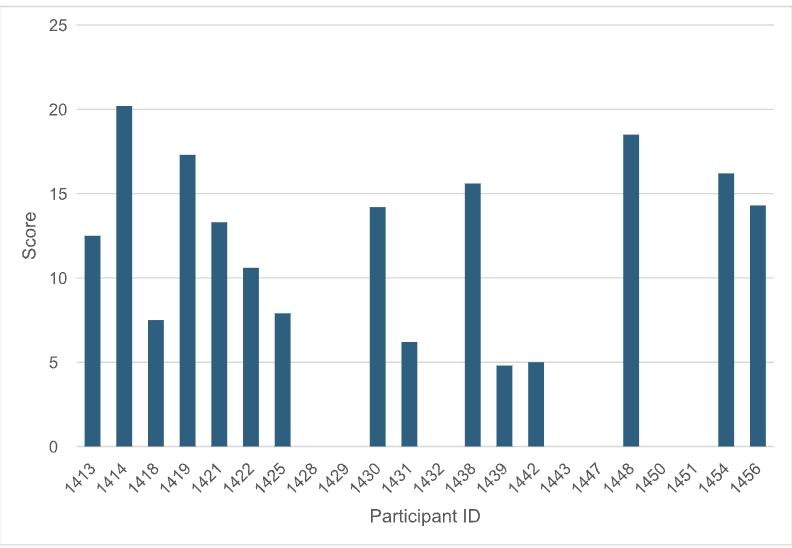
Figure 1: For each participant/method, the score of the best Round 1 molecule is plotted.
The aggregated score of all predicted molecules for each participant is another metric to compare the efficiency of the computational methods (Figure 2).
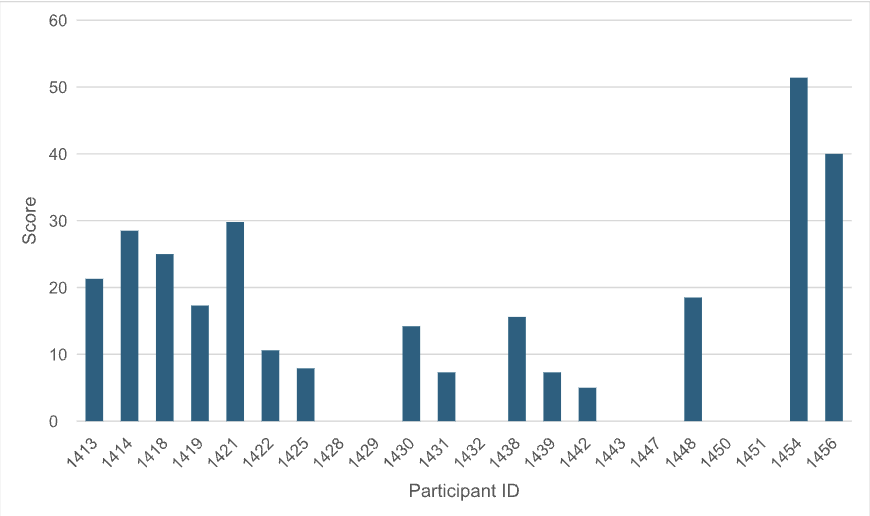
Figure 2: For each participant/method, the aggregated score of all Round 1 molecules is plotted
In another evaluation step, all participants were asked to predict hits out of the 1957 compounds selected in Round 1. The aggregated score of molecules predicted actives divided by the number of molecules predicted active is provided (Figure 3).
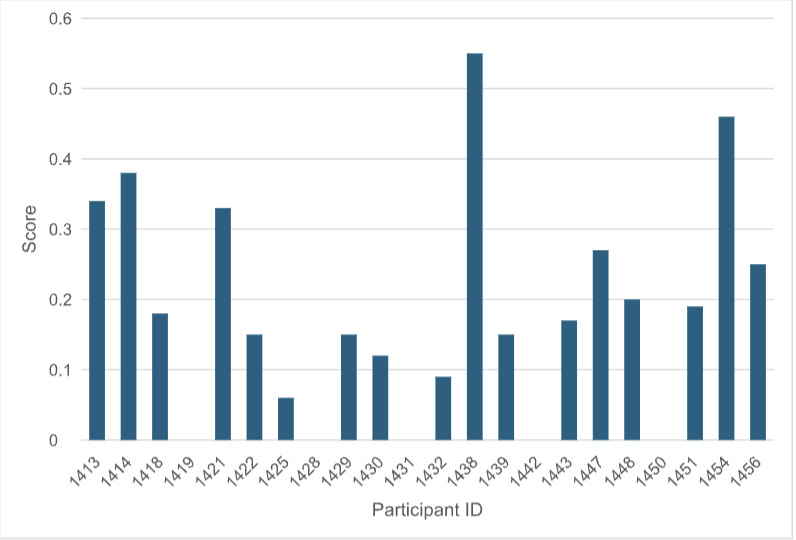
Figure 3: For each participant/method, the aggregated score of virtual screening hits predicted from the 1957 Round 1 compounds divided by the number of compounds predicted active is plotted.
The list of participants (de-anonymized with their permission), links to their computational methods, and associated scores are provided below.
| Participant Name | Links to computational methods | Score of best compound selected from Enamine REAL | Aggregated score of compounds selected from Enamine REAL | Aggregated score of predicted hits from the merged list of 1955 compounds from all participants / Number of predicted hits |
|---|---|---|---|---|
| Zhao, Bourne [U. of Virginia] | 1413 | 12.5 | 21.3 | 0.34 |
| Moretti, Scott, Meiler [Drug-it crowd-sourcing] | 1414 | 20.2 | 28.5 | 0.38 |
|
Moroz, Tarkhanova, Protopopov [Chem-space] |
1418 | 7.5 | 25 | 0.18 |
| Blay, McGibbon, Money-Kyrle, Houston [U. Edinburgh, Scotland] | 1419 | 17.3 | 17.3 | 0 |
| Kireev, Mettu, Wang [U. of Missouri] | 1421 | 13.3 | 29.8 | 0.33 |
| Zheng [Shanghai Jiao Tong Univ.], Lu, Jixian [Aureka Bio] | 1422 | 10.6 | 10.6 | 0.15 |
| 1425 | 7.9 | 7.9 | 0.06 | |
| Jensen [U. of Copenhagen, Denmark], Steinmann [Aalborg Univ. Denmark] | 1428 | 0 | 0 | 0 |
| 1429 | 0 | 0 | 0.15 | |
| 1430 | 14.2 | 14.2 | 0.12 | |
| Park, Durai [Korea Inst. of Science and Technology, Republic of Korea] | 1431 | 6.2 | 7.3 | 0 |
| Kandwal, Fayne [Dublin City Univ. Ireland] | 1432 | 0 | 0 | 0.09 |
| Cree, Pirie, Bieniek, Horton, Armstrong, Harris, Tatum, Cole [Newcastle Univ., U.K] | 1438 | 15.6 | 15.6 | 0.55 |
| Breznik, Liu, Denzinger, Pandit, Obendorf, Talagayev, Wolber [Free Univ. Berlin, Germany), Ricci (Univ. Messina, Italy] | 1439 | 4.8 | 7.3 | 0.15 |
| 1442 | 5 | 5 | 0 | |
| 1443 | 0 | 0 | 0.17 | |
|
Carosati [Univ. Trieste, Italy], Jukič [Univ. Maribor, Slovenia] |
1447 | 0 | 0 | 0.27 |
| Poda, Hoffer [Ontario Inst. for Cancer Res., Canada] |
1448 | 18.5 | 18.5 | 0.2 |
| 1450 | 0 | 0 | 0 | |
| Wells, Rosta [UCL, U.K.] | 1451 | 0 | 0 | 0.19 |
| Machado, Werhli, Schmitt [FURG, Rio Grande, Brazil] |
1454 | 16.3 | 51.4 | 0.46 |
| Koes, Dunn [U. of Pittsburgh] | 1456 | 14.3 | 40 | 0.25 |
NSP13 LIGANDS
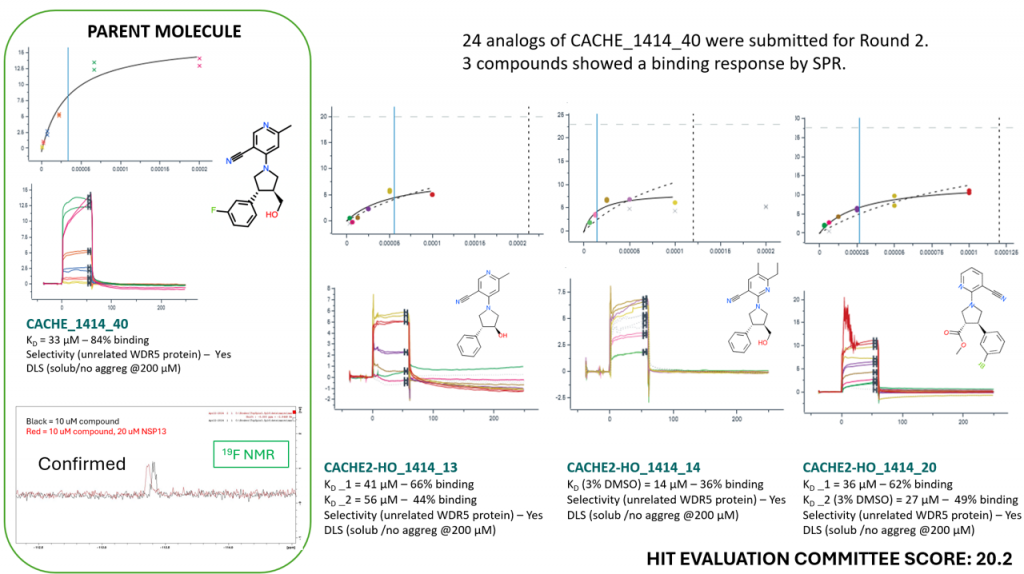
- To the best of our knowledge, no drug-like ligand with measurable KD was reported to date for SARS-CoV-2 NSP13.
- CACHE #2 participants collectively discovered over a dozen chemical series with measurable KD < 150 µM, some containing a fluorinated molecule with confirmed binding by 19F-NMR.
- Five of these have a score from the Hit Evaluation Committee above 15 and are more promising.
- These chemical starting points should be progressed into more potent molecules to investigate anti-viral activity.
- Two examples are shown below.
-------------------------------------------------------------------------------------------------------