Challenge #6
Click image to enlarge
Finding ligands targeting the triple Tudor domain of SETDB1
The sixth CACHE challenge is focused on SETDB1, a multi domain protein involved in epigenetic mechanisms and an immuno-oncology target.
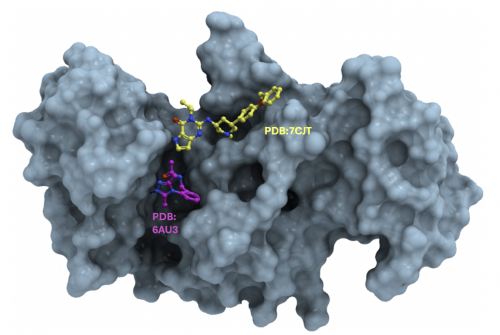
Participants are asked to find molecules occupying one or multiple subcavities of the histone binding groove of the SETDB1 triple Tudor domain (TTD). A few ligands (PDB codes 7CJT, 8UWP, 6AU3) occupy one or two of three subcavities that accommodate methylated or acetylated lysine sidechains, as shown in this structure overview. Read more details below.
CACHE is strongly committed to equity, diversity and inclusion. We strive to create an initiative that welcomes participants from countries, institutions and groups that are underrepresented in our research community. We endeavour to work with individuals who contribute to furthering the diversity of ideas and approaches, welcoming all communities to participate in our competitions.
All applications and submissions to this CACHE Challenge are subject to the CACHE Terms of Participation. Applications should be submitted using the online application form.
2024-05-08
2024-06-23
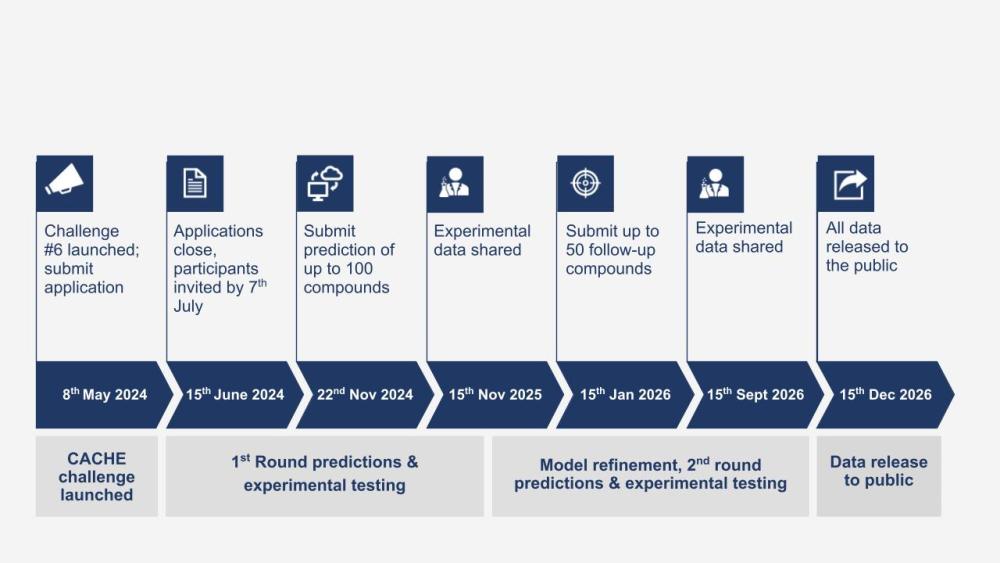
Timeline
Details
SETDB1 - Finding ligands targeting the triple Tudor domain
| Parameter | Description | |
|---|---|---|
| Disease association | SETDB1 suppresses tumor-intrinsic immunogenicity (Griffin et al. Nature 2021) and mediates resistance to immuno-therapy (Zhang et al. Nature 2021).
| |
| Challenge | Predict chemically novel ligands that bind the histone binding groove of the SETDB1 TTD.
| |
| Pharmacological landscape | The catalytic methyltransferase domain is atypical. No structure and no inhibitors were reported to date. A few compounds targeting the TTD of SETDB1 were reported that exploit one or two of three juxtaposed subcavities (PDB codes 7CJT, 8UWP, 6AU3) as described in this overview. Compounds targeting the TTD could be used as chemical handles for the development of future PROTACs inducing the degradation of SETDB1.
| |
| Macromolecular structures | Comparing structures in complex with different ligands reveals some structural plasticity at the binding site. One of the aromatic cages is sometimes obstructed and a gate between two of the three juxtaposed cavities was captured both in an open and closed state.
| |