Challenge #7
Click image to enlarge
Finding Selective Inhibitors of Phosphoglycerate Kinase 2
The seventh CACHE challenge is focused on PGK2, a kinase and a potential contraceptive drug target effecting sperm bioenergetics that is being actively pursued within the Gates Foundation non-hormonal contraceptive program. This challenge is supported by unpublished data provided by program members Damian Young and Kevin MacKenzie (BCM), Neelagandan Kamariah (inStem), Levon Halabelian and Hui Peng (SGC-Toronto), and Tim Willson (SGC-UNC).
CACHE participants are tasked with identifying orthosteric inhibitors that are selective for PGK2 over its close homologue PGK1, a glycolytic enzyme with available crystal structures in the PDB. All sidechains in the ligand binding pocket are conserved except for Y242 (PGK2) vs F242 (PGK1) and A255 (PGK2) vs T255 (PGK1) at the edge of the pocket.
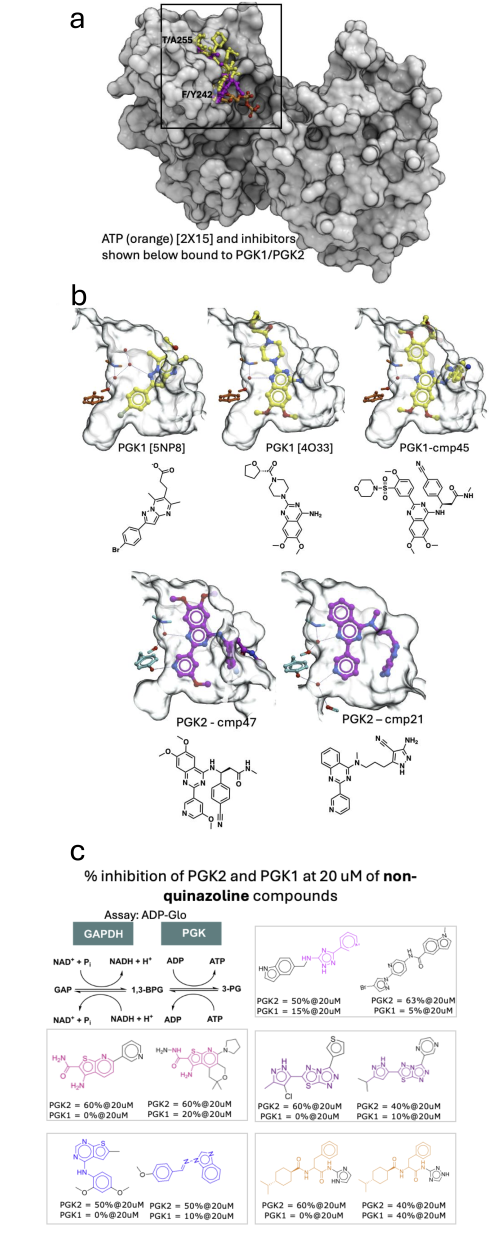
Figure 1: Structural chemistry of PGK1/2 inhibition. The ATP-bound structure has the PDB identifier 2X15 (a). Overview of structures (b) and ligands (c), most unpublished, that may inform the computational prediction of non-quinazoline inhibitors selective for PGK2 vs PGK1.
Available structural data include ATP-bound and ligand-bound hPGK1, ATP bound and apo mPGK2, and hPGK2 bound to two selective PGK2 quinazoline inhibitors (compound 21 and compound 47). hPGK1 bound to a selective quinazoline PGK1 inhibitor (compound 45) is also provided. Atomic coordinates and electron density files are available here.
Preliminary data on unpublished non-quinazoline ligands with some selectivity for PGK2 are shown (Figure 1c).
CACHE participants are also provided with 1388 hPGK2 hit candidates from a DNA-encoded library (DEL) screen (5 reads or more) that are not enriched in a hPGK1 screen. These compounds have not been tested off-DNA. As typical for DEL data, the signal-to-noise ratio is expected to be low and should be interpreted accordingly (ex: Montoya et al 2025).
The quinazoline series is well-advanced and will be published soon. CACHE #7 participants are therefore asked to discover SELECTIVE hPGK2 inhibitors that DO NOT contain a quinazoline.
CACHE will cover 100% of the costs of testing compounds experimentally. 100% of the costs of procuring compounds (either make-on-demand or custom-synthesized compounds) will be covered by participants. To lower the costs, CACHE will agree with Enamine on a preferred bulk price. CACHE will cover 50% of the costs for eligible Canadian academics and Canadian SMEs through a Strategic Innovation Fund awarded to Conscience by the Canadian government.
A maximum of 25 applicants will be invited to participate in this challenge, following a double-blind peer review process where each applicant is asked to review 5 applications.
CACHE is strongly committed to equity, diversity and inclusion. We strive to create an initiative that welcomes participants from countries, institutions and groups that are underrepresented in our research community. We endeavour to work with individuals who contribute to furthering the diversity of ideas and approaches, welcoming all communities to participate in our competitions.
All applications and submissions to this CACHE Challenge are subject to the CACHE Terms of Participation and must be submitted via the online application form.
2025-08-11
2025-10-09
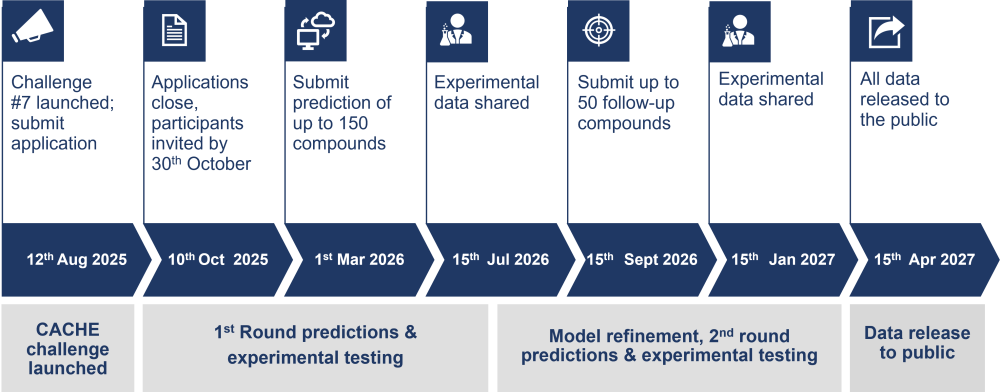
Timeline
Details
| Parameter | Description |
|---|---|
| Disease association | PGK2 catalyzes one of the two ATP producing reactions in the glycolytic pathway, is expressed exclusively during spermatogenesis and is essential for sperm function and male fertility in mice (Danshina et al. 2010). It is a potential contraceptive drug target effecting sperm bioenergetics (Liu et al 2016).
|
| Challenge | Predict orthosteric ligands that inhibit PGK2 with an IC50 < 10 µM, that are at least 20-fold selective for PGK2 over PGK1 and do not contain a quinazoline. Primary hits with an IC50 < 50 µM, that are at least 5-fold selective for PGK2 over PGK1 will be advanced to the hit expansion phase of the challenge.
|
| Pharmacological landscape | Several PGK1 and dual inhibitors have been reported (Chen et al. 2015, Bisland et al. 2019). The groups of Damian Young (BCM), Neelagandan Kamariah (inStem) and Halabelian/Peng/Willson (SGC) each identified quinazoline-based chemical series, using DEL (BCM), an ADP-Glo enzyme screen (inStem) and AS-MS (SGC). Non-quinazoline hits were also identified in the ADP-Glo screen. All compounds are selective for PGK2 over PGK1 and shared here before publication (Figure 1). |
| Macromolecular structures | Structures of hPGK1 and mPGK1 bound to ligands are available in the PDB (e.g., 2X15, 4O33, 4O3F, 5NP8), along with apo (2P9Q) and ATP-bound (2PAA) mPGK2. Additional structures include PGK2 bound to compounds 21 and 47, and compound 45 bound to PGK1. The adenine-binding cavity is enclosed and exploited by chemically distinct ligands. Only one sidechain central to the cavity and one at the edge differ between PGK2 and PGK1 (Figure 1a). Water molecules mediate key interactions. The remainder of the pocket is solvent-exposed, and one ligand shows low electron density in this region. All structures and electron densities are provided to CACHE #7 participants. |