Challenge #3
Click image to enlarge
Finding ligands targeting the macrodomain of SARS-CoV-2 Nsp3
The third CACHE challenge is focused on the macrodomain of SARS-CoV-2 Nsp3, a target with dozens of fragments and inhibitors in the PDB, open to both receptor-based and ligand-based approaches.
CACHE participants are asked to predict compounds with novel chemical templates without carboxylic acids and that compete with the substrate, ADP-ribose (ADPr). Read more under Details below.
This challenge is supported by the NIH's AViDD program.
Crystal structures are solved in James Fraser’s laboratory at UCSF.
CACHE is strongly committed to equity, diversity and inclusion. We strive to create an initiative that welcomes participants from countries, institutions and groups that are underrepresented in our research community. We endeavour to work with individuals who contribute to furthering the diversity of ideas and approaches, welcoming all communities to participate in our challenges.
All applications and submissions to this CACHE Challenge are subject to the CACHE Terms of Participation.
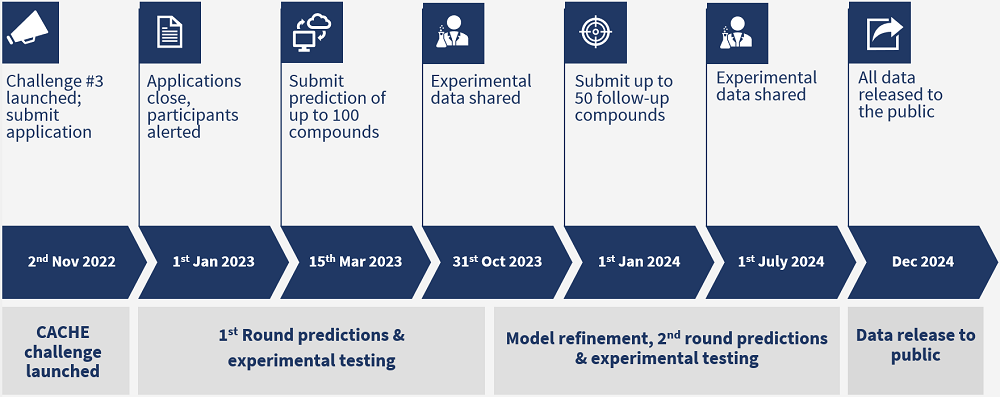
2022-11-02
2022-12-31
Timeline
Details
Macrodomain of SARS-CoV-2 Nsp3
| Parameter | Description | |
|---|---|---|
| Accession | YP_009725308.1 | |
| Disease association | Severe acute respiratory syndrome coronavirus 2 | |
| Challenge | To predict ligands that bind to the ADPr site of SARS-CoV-2 Nsp3 macrodomain (Mac1). | |
| Why the macrodomain? | Interferon response to coronavirus infection relies on ADP-ribosylation of target proteins. Nsp3 Mac1 removes this post-translational modification, thereby counteracting the immune response to viral infection (Russo et al. J Biol Chem 2021). Mutational analysis in cellular and animal models shows that Mac1 is a valid target against SARS-CoV (Kuri et al. J. Gen Virology 2011, Fehr et al. mBio 2016). | |
| Pharmacological landscape | A fragment screen by crystallography produced 54 crystal structures with fragments occupying the adenine binding cavity and 9 with fragments at the proximal ribose site (Schuller et al. Sci. Adv. 2021). Medicinal chemistry efforts led to inhibitors with low micromolar potency, and in some case cellular activity (Roy et al Antiviral Res 2022, Sherrill et al. Bioorg Med Chem 2022, Gahbauer et al. 2022 bioRxiv). Some of these data are summarized in James Fraser’s presentation at AViDD’s Open Science Forum (starting at 33’). | |
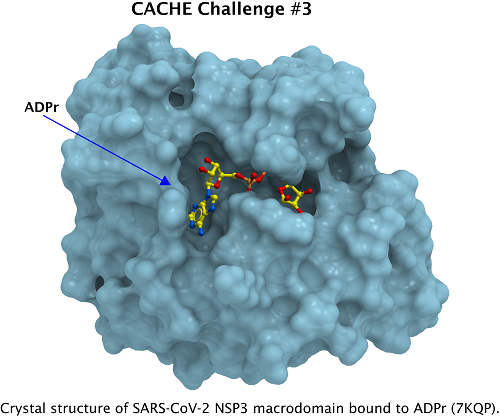
| Macromolecular structures | High-resolution structures of SARS-CoV-2 Nsp3 Mac1 are available in complex with its substrate, ADP-ribose ex: PDB 7BF5, 6Z6I, 6W02, 6Z5T, 7TX5, 7KQP (Figure 1) 7TWX, 6WOJ. Over fifty crystal structures with fragments occupying the adenine binding cavity and the proximal ribose site are available from the PDB and fragalysis (Schuller et al. Sci. Adv. 2021, Bajusz et al. Nat. Commun. 2021). Structures with multiple lead-like molecules are in the PDB with group deposition IDs G_1002236, G_1002238 and G_1002239. | |