WHAT IS CACHE?
The CACHE (Critical Assessment of Computation Hit-finding Experiments) Challenges offer an open competition platform to help accelerate one of the early stages of drug discovery. CACHE will help define the state-of-the-art in molecular design by providing unbiased, high quality experimental feedback on computational hit-finding predictions. CACHE drives discovery in areas of market failure, benchmarks the entire computational hit-finding workflow, and fosters collaboration among industry, SMEs, and academia.
In 2024, Conscience became the steward for the CACHE Challenges, which were originally launched by the SGC. Conscience believes that the CACHE challenges provide an excellent opportunity to drive forward their mission of addressing market failure in the current drug discovery system.
Subscribe to the Conscience mailing list to receive information about CACHE, including news, challenges and call for applications.
WHY PARTICIPATE IN CACHE ?
An important first step in the development of a small molecule drug is to screen large libraries of drug-like molecules against a given protein target, either experimentally or computationally.
These computational screening methods are poised to significantly and rapidly improve in coming years thanks to leaps in computational power, dramatic expansion of the accessible chemistry space, improvements of physics-based methods/force fields and maturation of deep learning.
CACHE will reveal the most efficient computational methods for hit-finding and guide future technological improvement.
CACHE builds on the power of crowd sourcing by attracting funding from industry, governments and foundations to support its infrastructure. In addition, challenge-specific funding mechanisms will give disease-focused funders the opportunity to recruit community-wide efforts to proteins of their interest.
An ancillary and valuable benefit of CACHE will be to discover ligands for new protein targets and thereby expand the open science paradigm for drug discovery.

HOW DOES CACHE WORK?
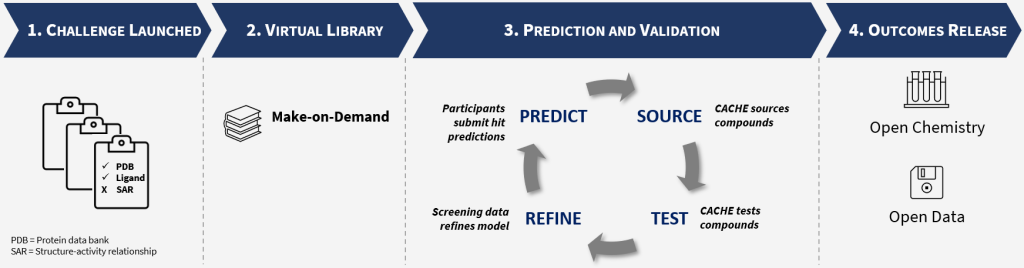
CACHE will initiate a new hit-finding benchmarking exercise every four months. These public competitions (challenges) will have the added benefit of identifying new chemical starting points for biologically interesting targets. Each competition will focus on a new protein target representing one of the following five possible challenges in hit-finding (Based on the type of target data available).
- Protein structure in complex with a small molecule, some structure-activity relationship (SAR)
- Protein structure in complex with a small molecule, no SAR
- Apo protein structure
- No experimentally determined protein structure, some SAR
- No experimentally determined protein structure, no SAR
For each challenge, participants will be invited to use their computational methods to find hits (drug-like molecules) for a protein target selected by CACHE. CACHE will procure the molecules from commercial vendors and test them experimentally with two rigorous binding assays. Compounds from de novo design methods will be procured by participants [details]. The purity and solubility of active molecules will also be evaluated experimentally. CACHE will provide each participant with the experimental protein binding data for their prediction. Using this knowledge, participants may predict a second round of molecules that will be experimentally tested by CACHE. Additionally, the structures of all selected molecules, but not their activity, will be made available to all participants who will be asked to use their computational method to identify hits. The final evaluation process will incorporate:
- experimental hit rate
- affinity and physico-chemical properties of the hits from a participant’s selection
- hit selection from the pooled library.
A panel of experienced medicinal chemists will also comment on the hits.
Participants will be asked to disclose their approaches in sufficient detail to enable an expert in the area to understand the methodology while computational algorithms can remain proprietary. At the end of the competition, all chemical structures and associated activity from all participants will be disclosed to the public. Top-performing participants and those who wish to will be de-anonymized.
In summary, Participants will use their computational method to predict hits that will be tested experimentally by CACHE. Each challenge will involve two cycles of predictions in order to give participants the opportunity to incorporate learnings from the first round into their designs. At the end of each challenge; following expert review, CACHE will release all data, including chemical structures, to the public.
ACCESS FEE
The access fee for entering a CACHE challenge and having compounds procured and tested varies depending on the challenge. Please refer to the specific Challenge number's page for further details. Canadian SMEs and academic participants can have up to 50% of their costs covered.
We encourage potential participants from academia to include these costs in their grant applications focused on the development and experimental validation of virtual screening methodologies. CACHE will support these applications, for example in the form of letter of support. Please contact us at [email protected] for details.
Disease foundations and other funders may in some cases sponsor challenges and cover the access fee for top candidates (based on the double-blind peer-review process taking place at the outset of each challenge).