Challenge #8
Click image to enlarge
Finding Ligands Targeting GID4
The eighth CACHE challenge is focused on GID4, the substrate-binding unit of the CTLH ubiquitin ligase complex. Compounds binding GID4 can lead to targeted protein degradation strategies that highjack CTLH E3 ligase activity for therapeutic applications across disease areas.
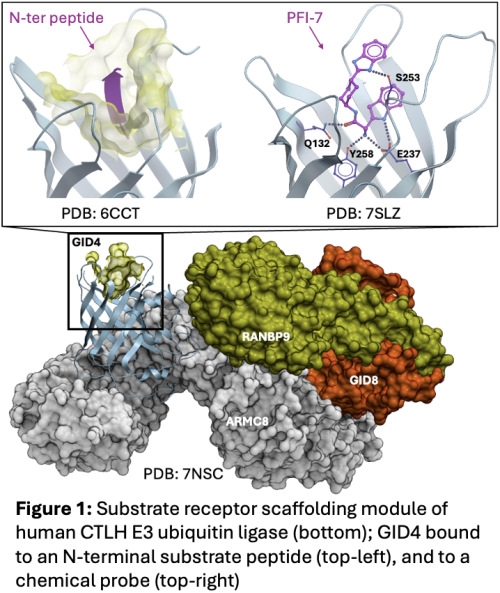
Participants are asked to find chemically novel molecules occupying the substrate binding pocket of GID4, occupied by a peptide in structure 6CCT and by a potent and selective chemical probe in structure 7S (see Figure)
CACHE will cover 100% of the costs of testing compounds experimentally. 100% of the costs of procuring compounds (either make-on-demand or custom-synthesized compounds) will be covered by participants. To lower the costs, CACHE will agree with Enamine on a preferred bulk price. CACHE will cover 50% of the costs for eligible Canadian academics and Canadian SMEs through a Strategic Innovation Fund awarded to Conscience by the Canadian government.
A maximum of 25 applicants will be invited to participate in this challenge based on a double-blind peer review (each applicant reviews 5 applications) and a rigorous evaluation process by the CACHE Application Review Committee.
CACHE is strongly committed to equity, diversity and inclusion. We strive to create an initiative that welcomes participants from countries, institutions and groups that are underrepresented in our research community. We endeavour to work with individuals who contribute to furthering the diversity of ideas and approaches, welcoming all communities to participate in our competitions.
All applications and submissions to this CACHE Challenge are subject to the CACHE Terms of Participation. Any questions pertaining to applications should be emailed to [email protected]
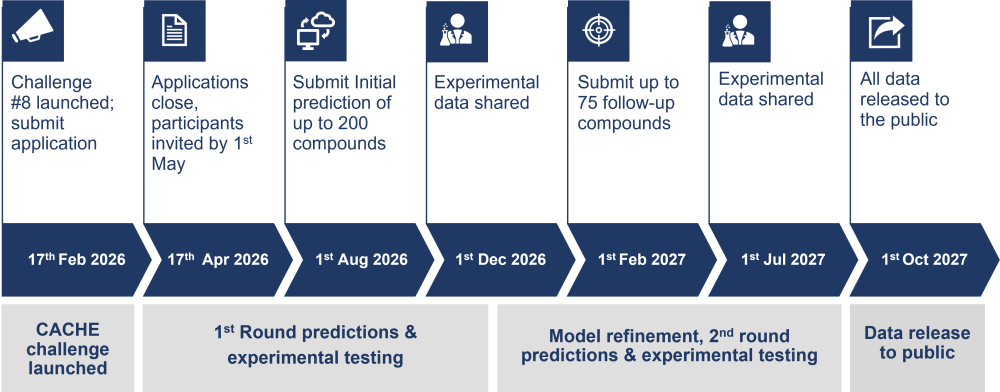
Timeline
Details
| Disease Association | Ligands binding GID4 can serve as chemical handles towards the elaboration of PROTACs, a pharmacological modality to induce the destruction of disease associated proteins by the endogenous protein degradation machinery |
|---|---|
| Challenge | predict chemically novel ligands that bind the substrate binding site of GID4. |
| Pharmacological landscape | 214 ligands were found from a Google Big Query (our thanks to Steve Boyer and Mike Moser from Molecular Assets for compiling this list). A few are in the PDB, including ligands 8V1P, 7SLZ, 7U3E, 7U3F, 7U3I, 7U3G, 7U3H, 7U3J, 7U3K, and PROTACS 8X7G and 8X7H." |
| Macromolecular structures | While the binding site is overall conformationally conserved across GID4 structures in the PDB, sidechain and backbone flexibility are observed, notably at loops Q132-Y139 and H275-Q282. |