Summary of CACHE#3 Round 1 experimental screening pipeline
CACHE#3 compounds are tested experimentally at the Structural Genomics Consortium, University of Toronto, with contributions from Suzanne Ackloo, Cheryl Arrowsmith, Albina Bolotokova, Irene Chau, Kristina Edfeldt, Pegah Ghiabi, Elisa Gibson, Oleksandra Herasymenko, Rachel Harding, Zarah Hejazi, Scott Houliston, Ashley Hutchinson, Carla Kharadjian, Peter Loppnau, Sumera Perveen, Matthieu Schapira and Madhushika Silva. Structural biology is conducted at UCSF, with contributions from Galen Correy and James Fraser.
THE CHALLENGE
- CACHE participants were asked to use their computational methods to find hits for the Macrodomain 1 of SARS-CoV-2 NSP3. See details.
- After a double-blind peer review where each applicant reviewed 5 applications, 23 participants joined the challenge, representing a diverse array of physics-based and AI computational methods.
- Participants collectively selected 1739 compounds (no more than 100 compounds per participant) that we ordered and received from Enamine.
- All experimental data except the structure of the compounds are provided here.
- Chemical structures will be revealed, CACHE participants de-anonymized, and computational methods ranked at the end of the challenge, upon completion of Round 2 (hit expansion).
All 1739 compounds were screened in two independent runs at 100 µM in a fluorescence-based HTRF competition assay as described in https://doi.org/10.1073/pnas.2212931120
An excellent correlation was observed between the two runs:
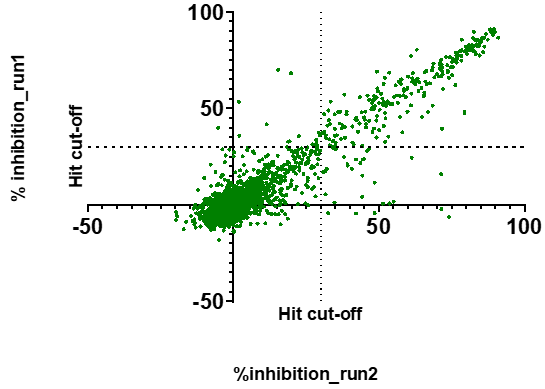
281 compounds showed >30% inhibition in at least one of the runs and were advanced to the next step. An additional 21 compounds showed inconclusive signal (overflow) in both runs and were also advanced to avoid false negatives.
The resulting 302 compounds were re-tested in the HTFR assay at 25, 50 and 100 µM in duplicate. 150 compounds showed clear dose dependent inhibition, >30% inhibition at 100 µM, no fluorescence interference and were advanced to the next step. The signal was ambiguous for another 16 compounds that were also advanced to the next step, to avoid false negatives.
The solubility and aggregation of 166 compounds was evaluated by DLS to determine the maximum concentration that could be used in the subsequent SPR dose response experiment.
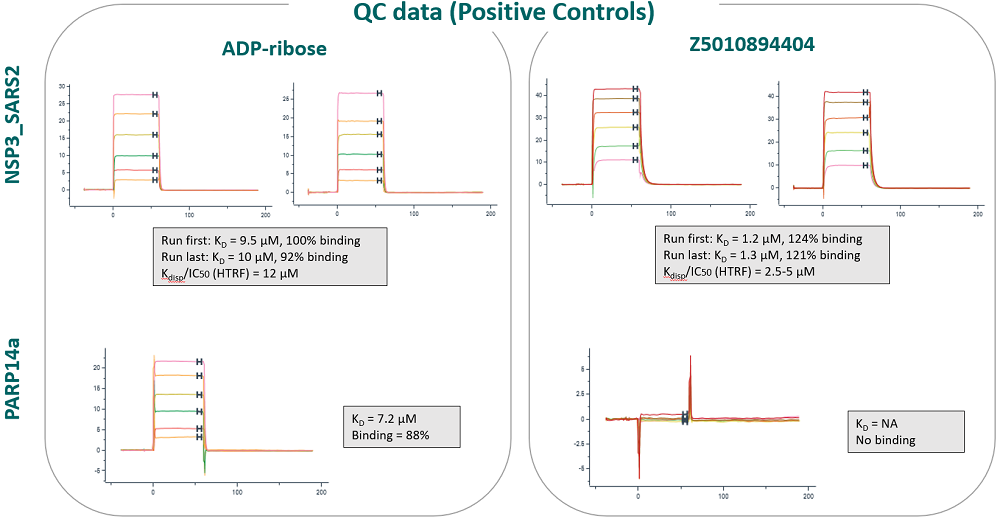
An SPR binding assay was validated with ADP-ribose and three positive controls, published in https://doi.org/10.1073/pnas.2212931120
Z5010894404 published Ki: 0.4 µM
Z5265428218 published Ki: 1.5 µM
Z5010894395 published Ki: 24 µM
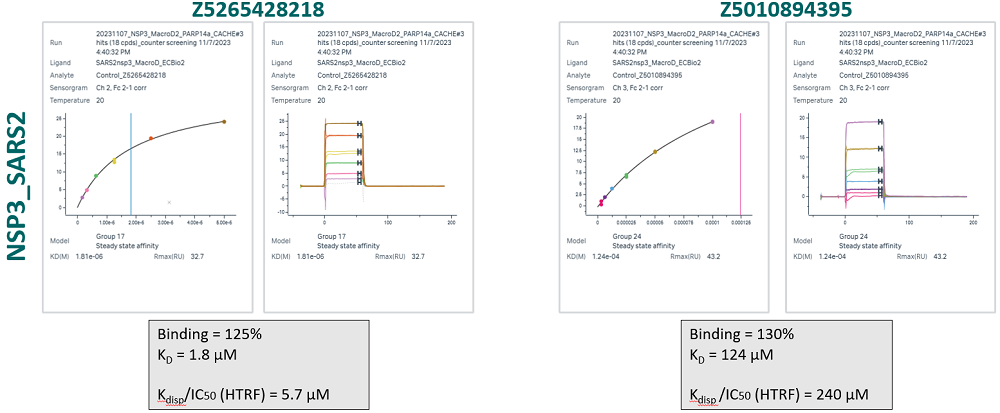
166 compounds were tested in a 6-point dose-response experiment by SPR. Top concentration was set to 100 µM, 50 µM or 30 µM, based on the solubility measured by DLS. 30 compounds had KD < 100 µM, > 30% of expected binding level, Chi2 < 10% Rmax and were advanced to the next step. Another 6 compounds had unclear data and were also advanced.
36 compounds were tested again by SPR, but with 4% DMSO instead of 2% DMSO to further increase the solubility of the compounds. Compounds were tested both against NSP3 (target) and PARP14 (anti-target), but PARP14 data was not used to dismiss compounds. 28 compounds selected by 9 participants were confirmed and advanced to Round 2: hit expansion. These 9 participants were asked to select 50 follow-up compounds to generate a convincing SAR around their experimental hits.
All screening data are available along with manuals to understand SPR and DLS data. Compound structures will be included once Round 2 (hit expansion) is complete.