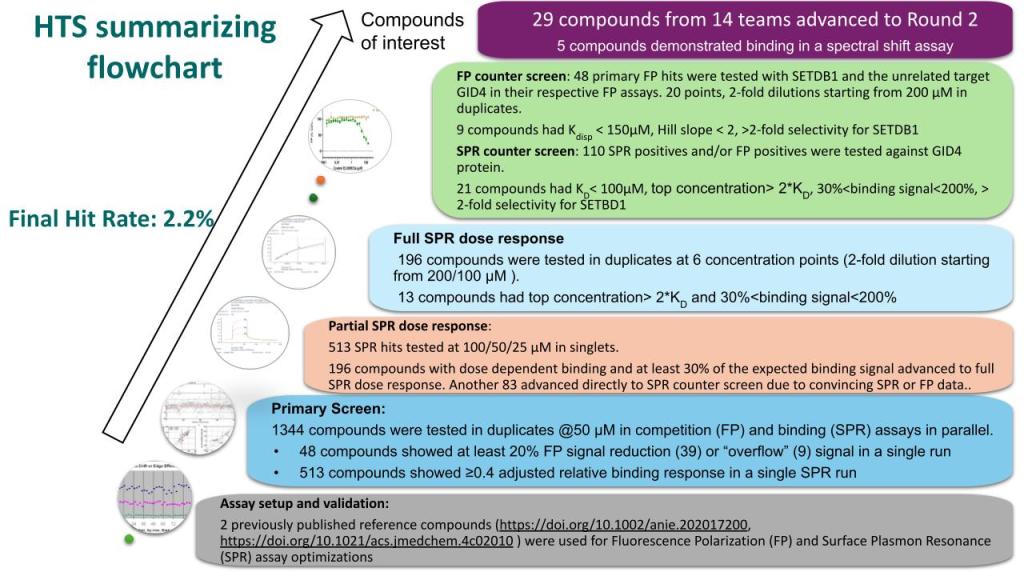
Summary of CACHE#6 Round 1 Experimental Screening Pipeline
CACHE#6 compounds are tested experimentally at the Structural Genomics Consortium, University of Toronto, with contributions from Oleksandra Herasymenko, Madhushika Silva, Elisa Gibson, Albina Bolotokova, Peter Loppnau, Rachel Harding, Ashley Hutchinson, and Matthieu Schapira
Here are the results from the experimental validation of compounds from the first round (hit identification) of CACHE challenge #6. The full data will be released upon completion of the second round (hit expansion).
THE CHALLENGE
- CACHE#6 participants were asked to use their computational methods to find hits for the triple tudor domain of SETDB1. See details.
- After a double-blind peer review where each applicant reviewed 5 applications, 18 participants joined the challenge, representing a diversity of physics-based and AI computational methods.
- Participants collectively selected 1344 compounds (no more than 100 compounds per participant) that we ordered and received from Enamine.
- All experimental data except the structure of the compounds are provided here.
- Chemical structures will be revealed, CACHE #6 participants de-anonymized, and computational methods ranked at the end of the challenge, upon completion of Round 2 (hit expansion).
SUMMARY OF THE EXPERIMENTAL PIPELINE
SUMMARY OF EXPERIMENTAL DATA AND ASSOCIATED COMPUTATIONAL METHODS
Method | Compound | Binds in SPR assay | Competes in FP Assay | Binds in Spectral Shift Assay | Confidence that the hit is real |
| 2330 | CACHE6-HI_2330_51 | yes |
|
| medium |
| 2334 | CACHE6-HI_2334_23 | yes |
|
| medium |
| 2336 | CACHE6-HI_2336_77 | yes |
|
| medium |
| 2336 | CACHE6-HI_2336_8 | yes |
| yes | medium |
| 2336 | CACHE6-HI_2336_85 | yes |
|
| medium |
| 2337 | CACHE6-HI_2337_64 | yes |
|
| medium |
| 2356 | CACHE6-HI_2356_111 | yes |
|
| medium |
| 2356 | CACHE6-HI_2356_26 | yes |
|
| medium |
| 2356 | CACHE6-HI_2356_40 | yes |
|
| medium |
| 2357 | CACHE6-HI_2357_38 | yes |
| yes | medium |
| 2357 | CACHE6-HI_2357_60 | yes |
|
| medium |
| 2357 | CACHE6-HI_2357_69 | yes |
| weak | medium |
| 2363 | CACHE6-HI_2363_66 | yes |
|
| medium |
| 2366 | CACHE6-HI_2366_31 | yes |
|
| medium |
| 2366 | CACHE6-HI_2366_43 | yes | yes | yes | high |
| 2369 | CACHE6-HI_2369_57 | yes |
|
| medium |
| 2375 | CACHE6-HI_2375_11 | yes |
|
| medium |
| 2375 | CACHE6-HI_2375_46 | yes |
|
| medium |
| 2377 | CACHE6-HI_2377_25 | yes |
|
| medium |
| 2379 | CACHE6-HI_2379_11 | yes |
|
| medium |
| 2486 | CACHE6-HI_2486_40 | yes |
|
| medium |
| 2336 | CACHE6-HI_2336_58 | yes |
| low | |
| 2336 | CACHE6-HI_2336_81 | yes |
| low | |
| 2376 | CACHE6-HI_2376_12 | yes |
| low | |
| 2376 | CACHE6-HI_2376_21 | yes |
| low | |
| 2376 | CACHE6-HI_2376_27 | yes |
| low | |
| 2376 | CACHE6-HI_2376_34 | yes |
| low | |
| 2376 | CACHE6-HI_2376_72 | yes |
| low | |
| 2376 | CACHE6-HI_2376_76 |
| yes | weak | medium |
VALIDATION OF EXPERIMENTAL ASSAYS
Known SETDB1 binders were used as positive controls to validate the fluorescence polarization (FP) assay:
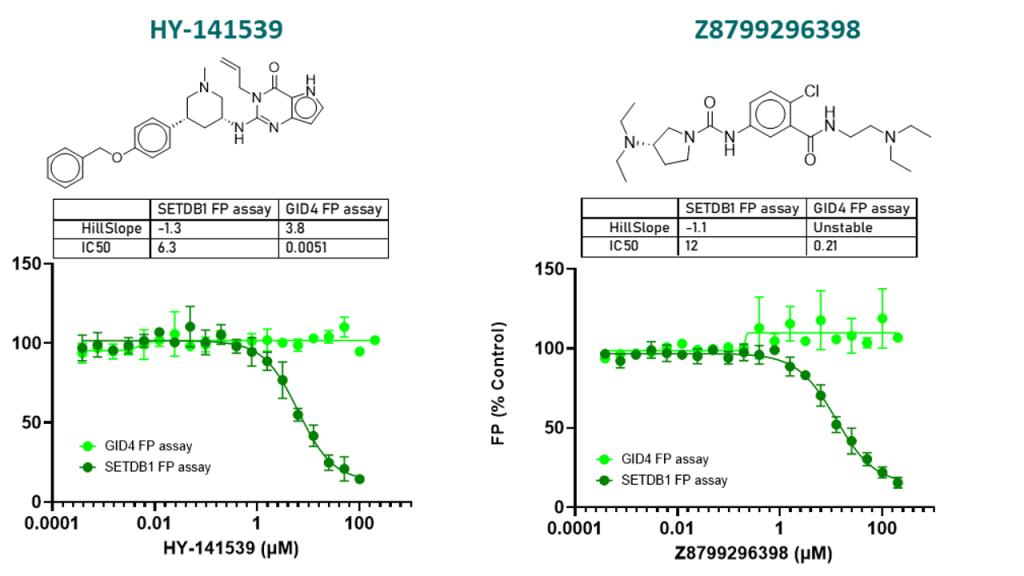
FP assay conditions: recombinant SETDB1 [construct 197-403aa with C-terminal His and N-terminal Avi tagged] at 2.5 µM; FITC labelled H3K9me2K14ac (1-25) at 10 nM; Buffer: 20 mM HEPES pH 7.5, 50 mM NaCl, 5 mM MgCl2, 0.01% (v/v) Triton X-100, 2 mM DTT, 4% (v/v) DMSO at room temperature.
The same compounds were used as positive controls to validate the surface plasmon resonance (SPR) assay:
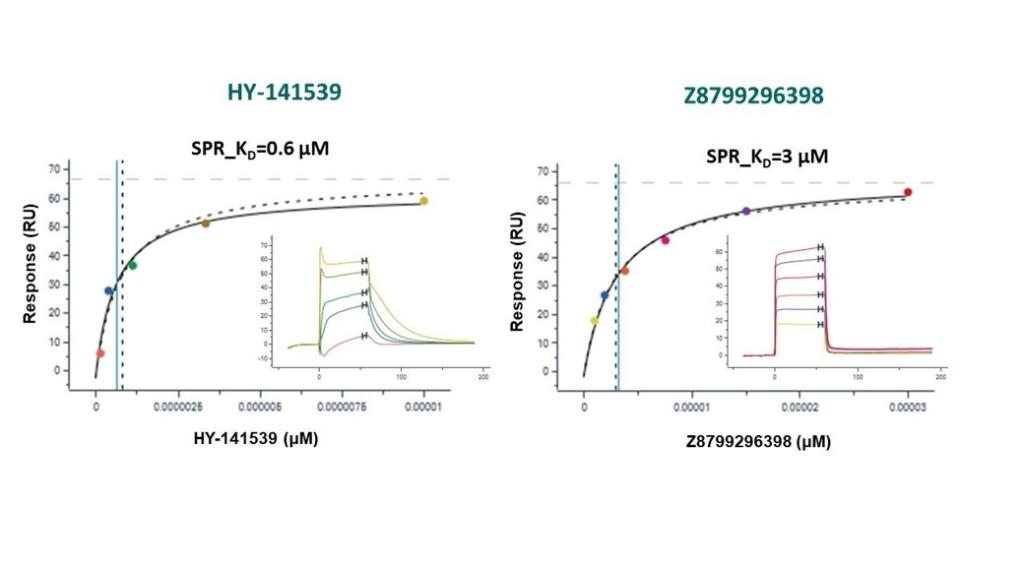
SPR assay conditions: recombinant SETDB1 [construct 197-403aa with C-terminal His and N-terminal Avi tagged] was immobilized onto the active flow cells (flow cell 2) of 8 channels on a SA sensor chip with buffer 10mM HEPES pH 7.4, 150mM NaCl, 0.005% v/v Tween 20, 0.2% w/v PEG 3350, 0.5 mM TCEP and 4% (v/v) DMSO yielding around 3000-4000 RU at 20 0C. Method – Multicycle kinetics.
The same compounds were used as positive controls to validate the spectral shift binding assay:
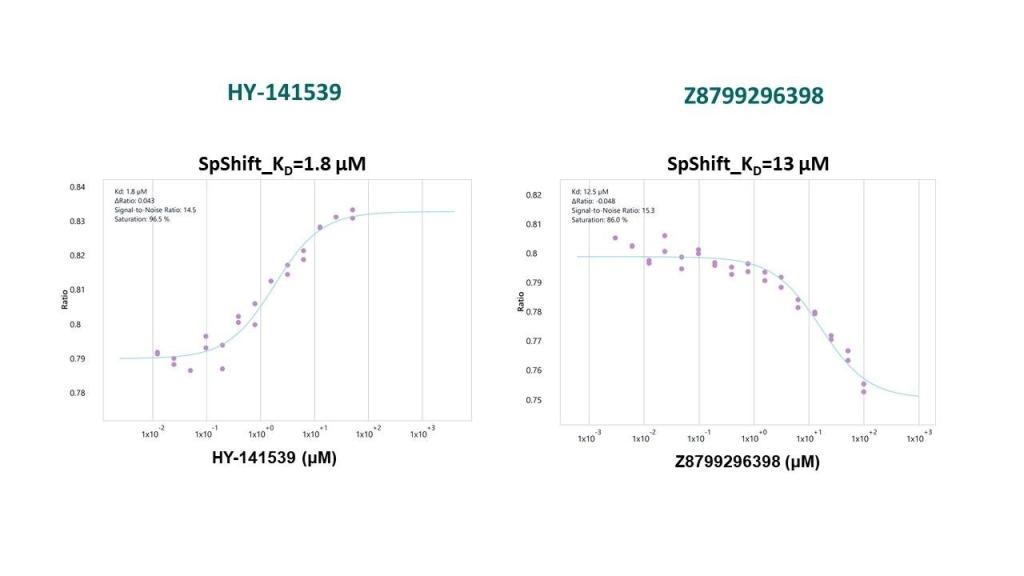
Spectral shift assay conditions: recombinant SETDB1 [construct 197-403aa with C-terminal His and N-terminal Avi tagged] labelled via His-tag with SpShift-optimized fluorescent dye at 25 nM. Buffer: 50 mM HEPES pH 7.5, 150 mM NaCl, 0.005% Tween, 0.5 mM TCEP, 4% (v/v) DMSO at room temperature.